One of the more unpredictable parts of crystallography is also one of the most fundamental – will the protein crystallise? Despite decades of work done on protein crystallisation, and numerous ‘tricks of the trade’ to maximise the chances, whether a protein will actually form a crystal is still relatively random. With ALK2 we’ve crystallised it enough before to have optimised which bits of the protein to use to make it most stable and remove any flexible ends that might prevent crystallisation – a shorter construct without the GS loop if it’s on its own or a slightly longer construct with the GS loop if it is in complex with FKBP12. However even knowing that these proteins will crystallise, doesn’t mean that they will crystallise when you introduce a new compound that can alter the dynamics of the drop.
In my last post I set up several plates with 6 different compounds and got some crystal hits – the next step is to take those crystals and ‘mount’ them in small nylon loops (which is like going fishing under a microscope and is exactly as fiddly as it sounds), flash freeze them in liquid nitrogen and then send them on a trip to the diamond light source to see if we can get any useful data from them. All these crystals were grown at 4°C and so all mounting was also done at 4°C.
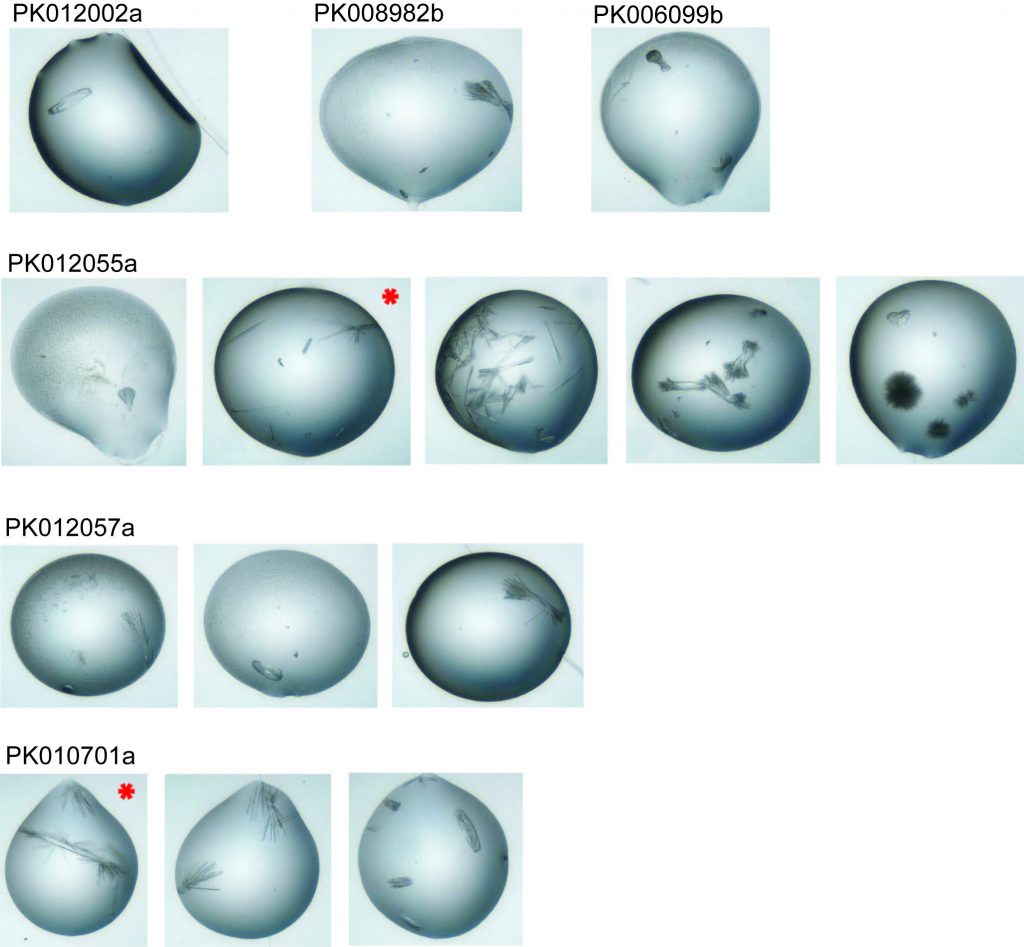
Drops from which crystals were mounted – the two with red stars diffracted well enough to give data sets.
These are the crystals mounted for each compound – as you can see they are quite narrow and needle-like. This can make them difficult to mount without breaking them or without getting multiple crystals in the loop which can then give overlapping and confusing diffraction patterns.
I managed to mount something from each well and sent them along on the next synchrotron trip to see if any useful diffraction data could be got from them.
Data Collection:
The synchrotron (The Diamond Light Source) has a number of different x-ray beamlines – this trip was to beamline I03. I wasn’t able to make it to the trip its self but trip leader Joe Newman collected some data for me. (Thanks Joe!)
Of the 16 crystals sent, several diffracted but only two gave decent enough data to collect a full dataset. Several of the others diffracted to a high resolution but there was more than one crystal in the loop (either due to the fact the crystals were very thin and needle like so hard to mount, or because several crystals had grown together) meaning that the data from each of the crystal fragments overlapped too badly to collect data separately.
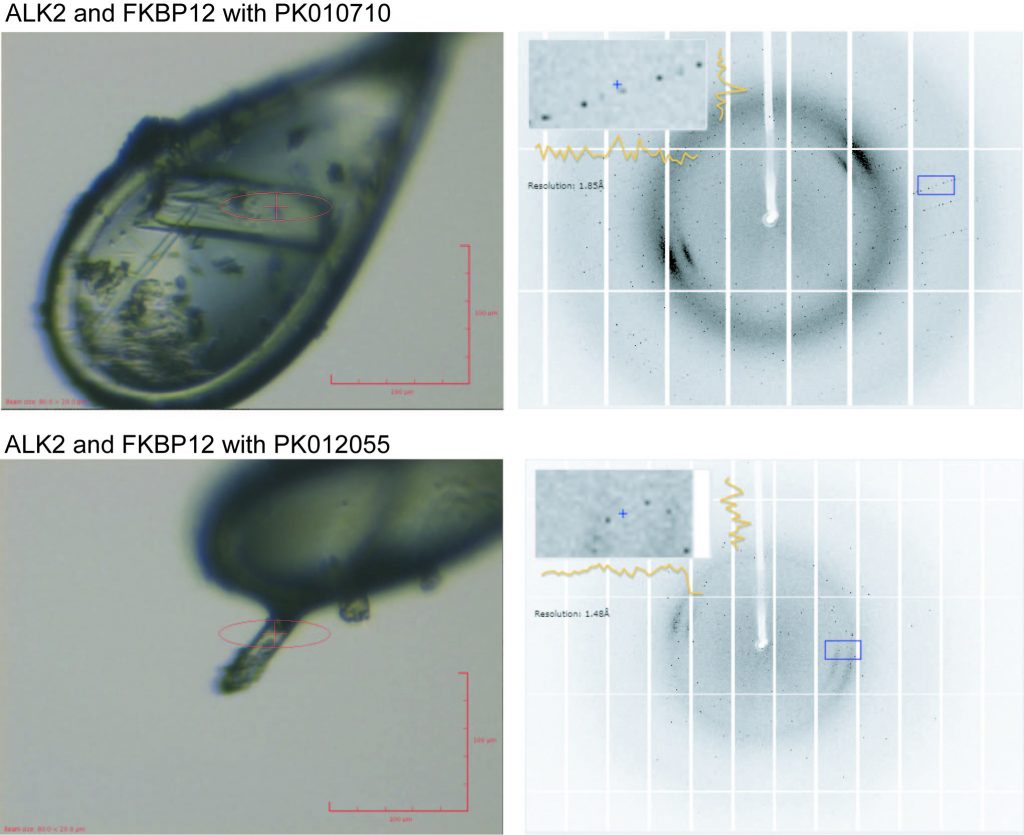
Shots of the two crystals that gave complete datasets: Left: mounted in the loops and right: the diffraction data collected from them.
The two data sets collected were for compounds PK010701 and PK012055 at a resolution of 1.6Å and 1.8Å respectively. It’s going to take a little while to process the data to actually see the protein with the ligand bound but this is a good start!
I’m going to try and mount some more crystals of the other compounds to see if I can get any in isolation rather than overlapping too badly.
For full experimental details you can find the zenodo link here.
