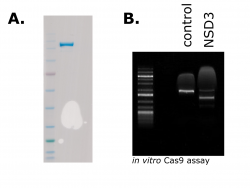
Bio NSD3 PWWP1 Expression and Purification Protocol
The NSD protein family has been implicated in cancers, Therefore they are considered as potential targets for cancer drug discovery. Here is the Expression and purification protocol of recombinant BIO NSD3 PWWP1 for further research. https://zenodo.org/record/4088451#.X4cd6WhKi70