Hi there! The last month of my life was taken over by making sure my PhD first year report was beautifully polished, but I have returned with results of a small compound screen:
These are all compounds that Jong Fu has already tested with his assays so we know they effectively inhibit ACVR1, but I thought it would be a good idea to try them out on my DIPG cells and see how well they can kill them. I included a few different variables in this screen to test what could make a difference to the outcome:
- I tested cells grown in 2D and 3D because this has been noted to change the outcome of compound screens in other systems
- I tested the compounds alone, or in addition to radiation in order to test whether the compounds were effective in addition to current DIPG treatments.
I tested how effective each compound is by growing cells for 7 days in the presence of the compound. At the end I measured how alive or healthy the cells were (i.e. their viability) by using a pre-developed kit that measures the amount of ATP inside the cell by using an enzyme that can produce light when ATP is present. ATP is measured because this is sometimes called the ‘energy currency of the cell’ so living cells have to have ATP inside them, whereas dead cells won’t and won’t be counted.
This screen produced a lot of data at once which is best visualised as heatmaps like these:
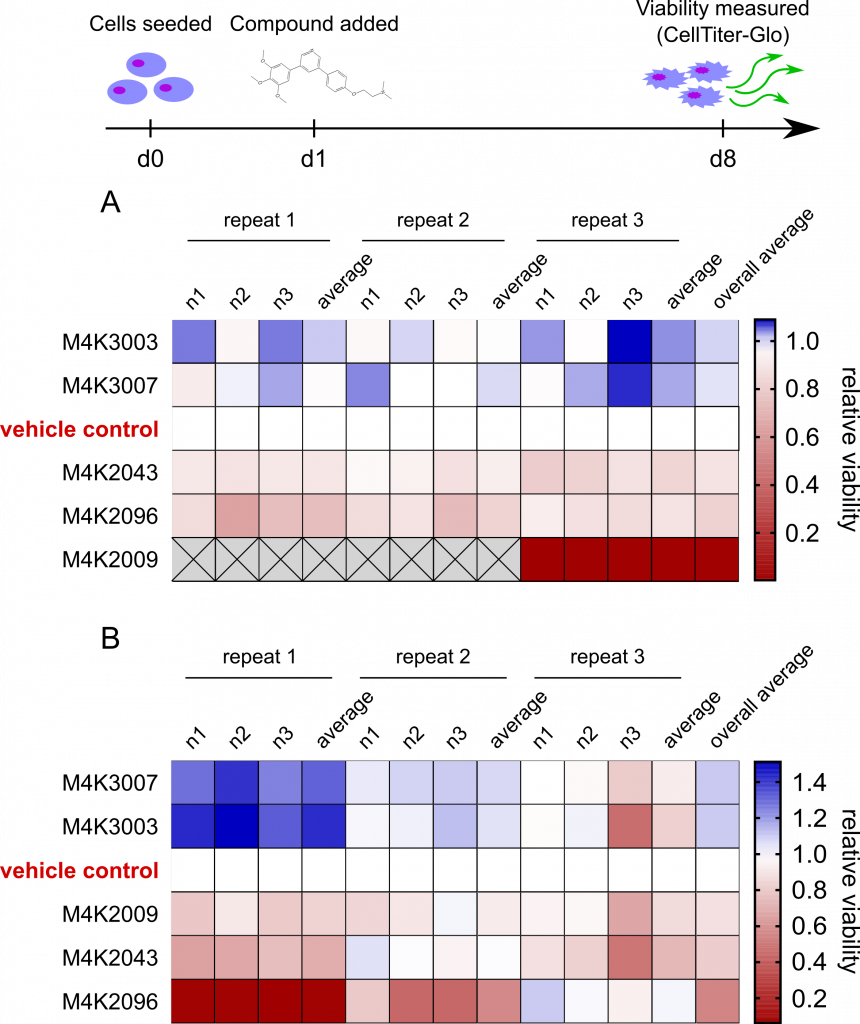
ACVR1 inhibitor M4K2009 is best at reducing mutant ACVR1 DIPG cell viability, but is not effective in ACVR1 wild-type cells
Heatmaps showing the viability (relative to an untreated control) of a 2D culture of mutant ACVR1 DIPG cells (A) or wild-type ACVR1 DIPG cells (B) following 7 days in the presence of 1μM of the indicated ALK2 inhibitors. The colour of each box indicates how viable the cells were at the end (where blue indicates an improvement in viability and red indicates a drop in viability). Each column indicates an individual (technical repeats, n1-3) grouped into those done at the same time (biological repeats 1-3).
From that heatmap you can see that M4K2009 is the best at killing ACVR1 mutant cells (the top heatmap), but it isn’t as effective in cells where the ACVR1 gene is not mutated (“wild-type” cells). In those cells instead M4K2096 is the most effective compound (but still not to the same extent as M4K2009).
But this is only the data for cells grown in 2 dimensions without a radiation treatment. If you want to see the full dataset, you can read my Zenodo post!
