It has now been almost 2 years since I set out to try and make fragments of the huntingtin protein which might be amenable to structural analysis with X-ray crystallography. X-ray crystallography is a fantastic method and allows us to see the molecules in very fine atomic detail which is important if we are to understand the intricacies of the elusive huntingtin protein molecule. It has been a long hard road with almost none of our extensive cloning efforts producing any expression constructs which made sufficient yields of protein. X-ray crystallography is a protein-expensive method so we need milligram (lots of protein). But finally, I have purified milligrams of different huntingtin fragment protein samples!!!
It should be made very clear that this has been a big team effort with cloner extraordinaire, Peter Loppnau, the eukaryotic production, Ashley Hutchinson and Alma Seitova as well as Linda Lin doing a lot of the heavy lifting on our cloning and eukaryotic production pipeline so I am very grateful for all of their hard work. Turns out that my construct design was fine but the expression vector we used made a huge difference (this is the piece of DNA which we insert different parts of the huntingtin gene). Switching from pFBOH-MHL to pBMDEL gave us great yields! Here is the C-terminal HEAT domain protein I purified – so much pure protein! All of the other data can be found on Zenodo.
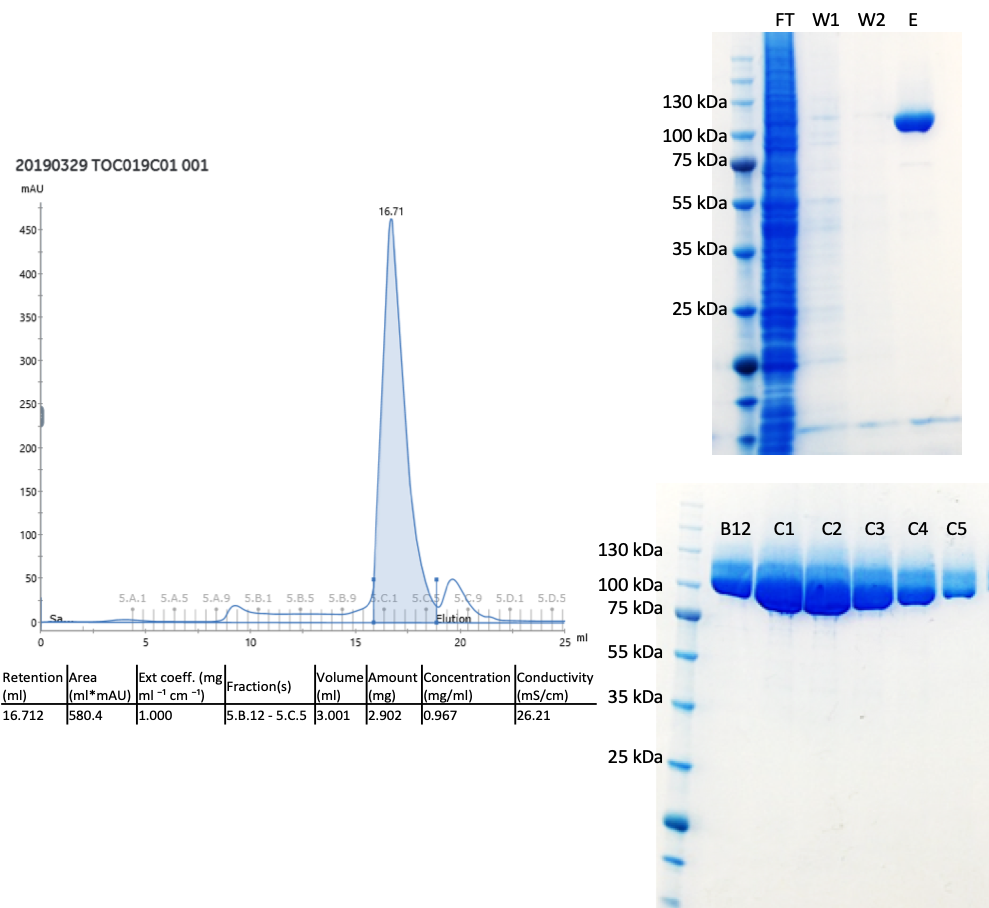
The gel filtration fractions were run on the gel before I had measured protein concentration – there is too much sample on the gel, but it looks great!
I have also been working to make the cleanest possible sample of the HTT-HAP40 protein complex – all data are on Zenodo. Having very clean samples should reduce prep-to-prep variation and reduce contamination of pesky proteins which are not either HTT or HAP40 in the sample. However, there is a trade off; more purification steps are needed to make a very pure sample but for every additional step the yield will decrease so I end up with less final material. However, I am very pleased with this sample (tempted to say cleanest prep ever?!).
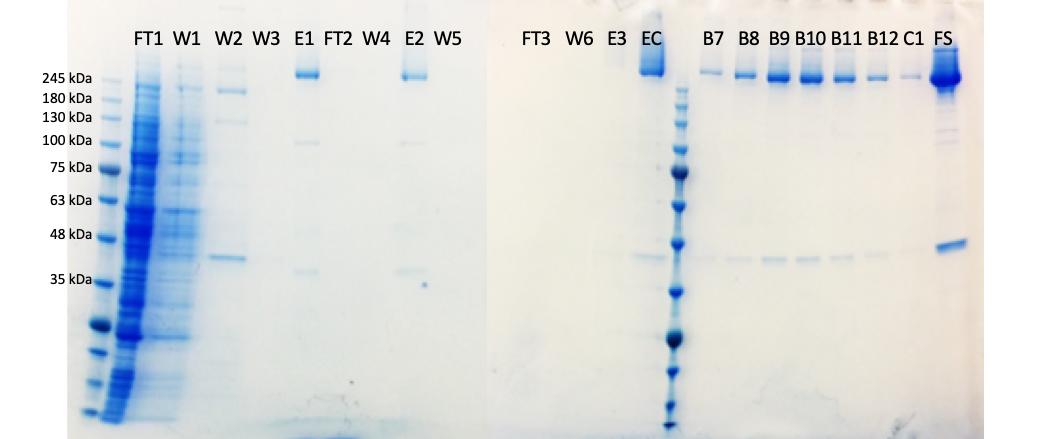
Here you can see fractions taken throughout the HTT-HAP40 prep from starting material on the left to the final sample (FS) on the right.
