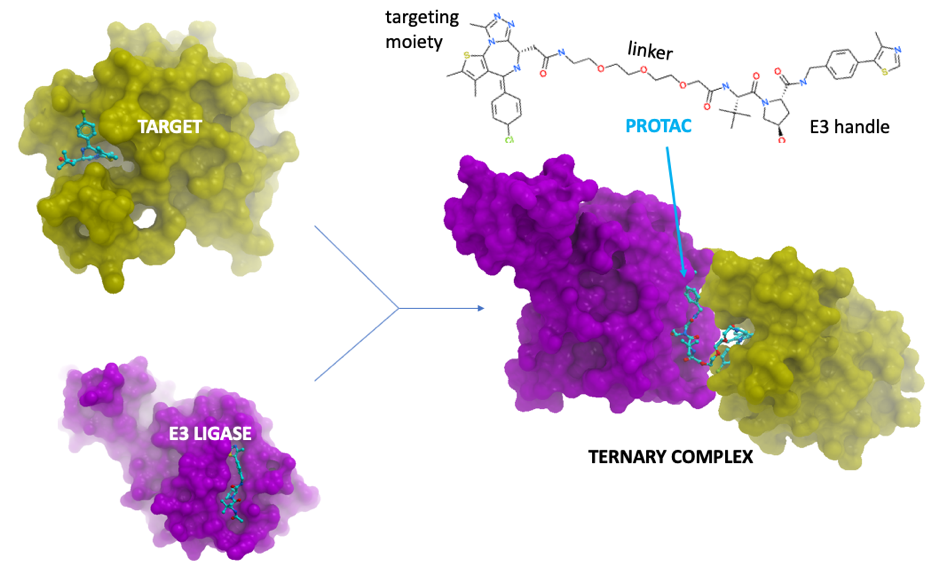
Proteolysis Targeting Chimeras (PROTACs) are small molecules that induce the degradation of their target and have shown considerable promise as a novel therapeutic modality.1-3 PROTACs are heterobifunctional compounds containing one chemical moiety that binds to the target protein of interest and a chemical handle that binds to an E3 ubiquitin ligase (Figure 1). Recruitment of the E3 ligase leads to ubiquitination and subsequent degradation of the target by the proteasome. The design of PROTACs remains an empirical process, likely because of an insufficient understanding of the molecular basis for target recruitment and formation of a ternary complex.4,5 Not all ligases are compatible with all targets, and linker chemistry and attachment points play a crucial role but are poorly understood.
 .
.
Figure 1: a PROTAC induces the formation of a ternary complex.
The goal of this research project is to test whether computational techniques can be used to rationally guide the design of PROTACs. I will be asking four questions:
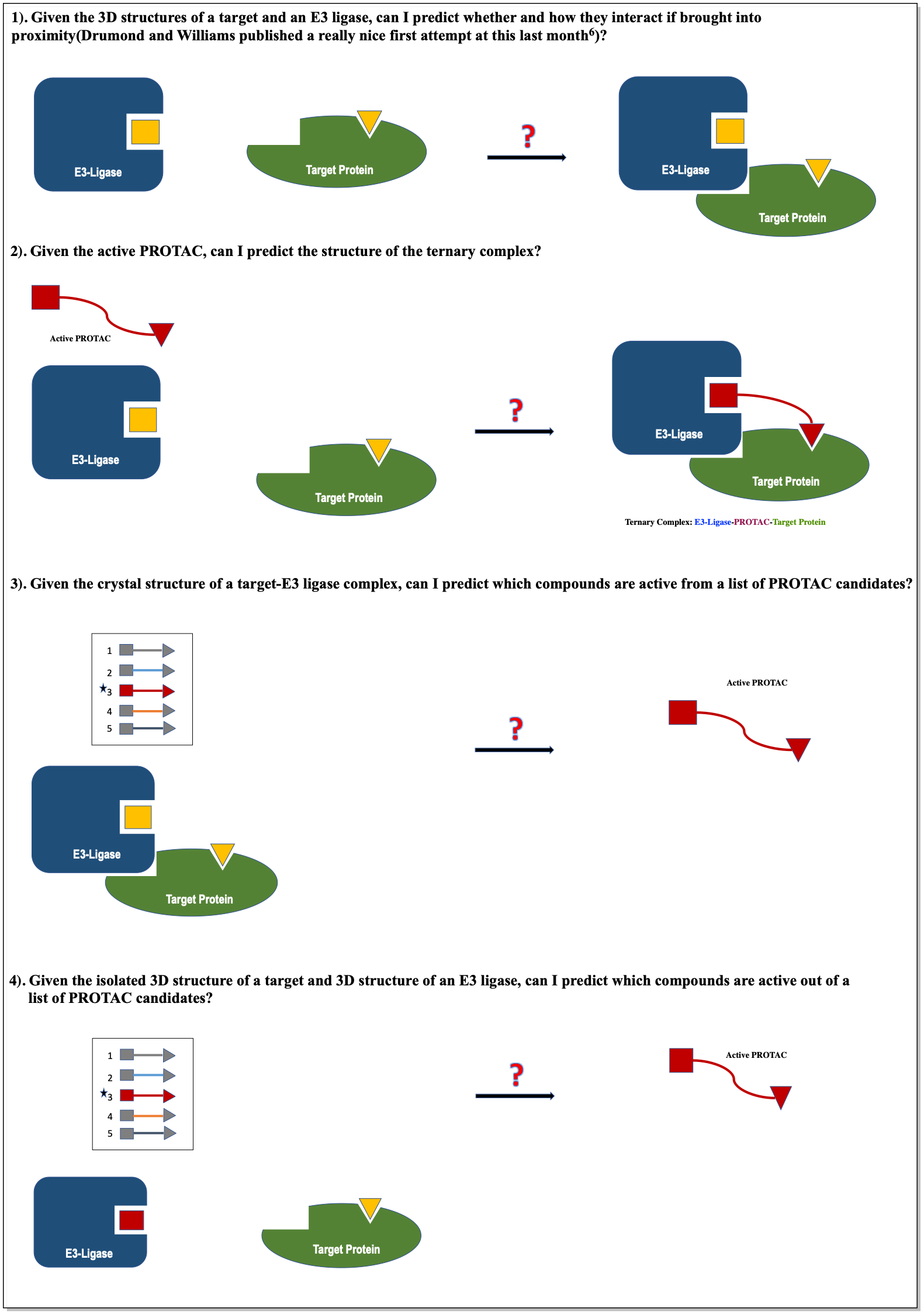
Virtual screening and structure-based drug design are quite unreliable techniques for drug-like molecules but are still used because they are generally performing better than random. Considering the structural complexity of the ternary complex central to PROTAC activity, and the chemical complexity of PROTAC molecules, in silico design of PROTACs seems even more challenging. Can rational approaches accelerate PROTAC discovery? I will try to systematically address this important question.
Reference:
- Crews, C. M. et al. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug. Discov., 16, 101-114 (2017).
- Scudellari, M. et al. Protein-slaying drugs could be the next blockbuster therapies. Nature., 567, 298-300 (2019).
- Churcher, I. Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones? J. Med. Chem., 61, 444-452 (2018).
- Fischer, E. S. et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol., 14, 706–714 (2018).
- Ciulli, A. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol., 13, 514–521 (2017).
- Williams, C. I. et al. In Silico modeling of PROTAC-medicated ternary complexes: Validation and application. J. Chem. Inf. Model. Doi:1021/acs.jcim.8b00872
