SARS-CoV-2 is a positive-strand RNA virus, depending on its multi-subunit machinery to replicate its RNA. This machinery is known as RNA-dependent RNA polymerase (RdRp). The catalytic subunit of RdRp, which is the core component of this machinery, is called nsp12. Nsp12 has little catalytic activity on its own and relies on accessory subunits to have full activity. These accessory factors, nsp7 and nsp8, increase RdRp template binding and processivity. (Yin et al., 2020)
RdRp is a well-established target for a class of antiviral drugs known as nucleotide analogs. Remdesivir is an example of such a group of drugs that inhibits RdRp. Remdesivir has been authorized to be used for special COVID-19 cases with severe symptoms in Canada. (https://healthycanadians.gc.ca/recall-alert-rappel-avis/hc-sc/2020/73621a-eng.php)
We previously analyzed the remdesivir-binding site of RdRp. We then shifted our focus on another druggable pocket of the RdRp complex, an allosteric site on the RdRp complex, which is this post’s topic.
In figure 1, we show the RdRp complex and highlight the site of interest, which is lined by amino acid residue M666 and which we refer to here as the M666 pocket. This site has a druggability score of 1.028 based on our results from Sitemap (Schrodinger, NY), indicating a druggable site (figure 2, Zenodo report).
M666 site is at the nsp8 binding site of nsp12. A compound binding this site with high potency may antagonize the formation of an active RdRp complex. It may inhibit the RdRp function, which is necessary for viral replication.
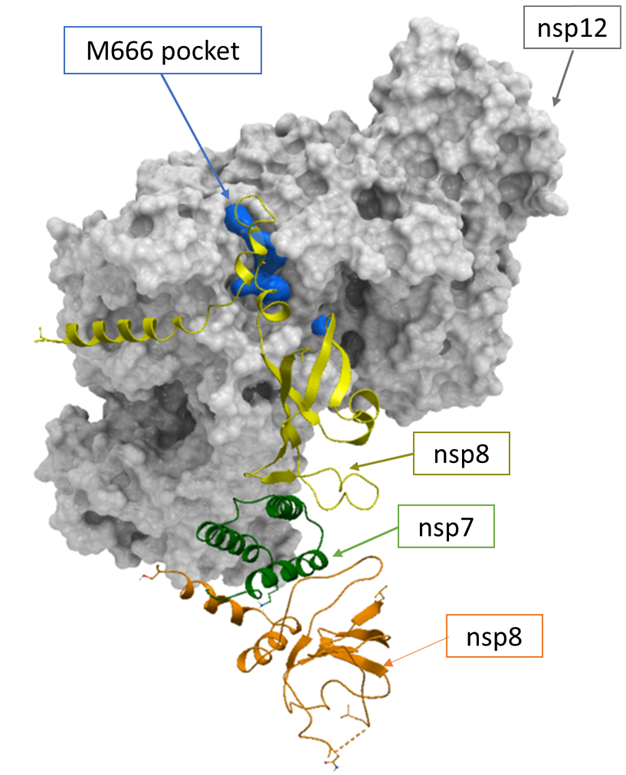
Figure 1. The apo nsp12-nsp7-nsp8 RdRp complex and the positioning of the M666 pocket at the interface of nsp12-nsp8.
We then selected the residues surrounding this pocket in proximity to bound nsp8. The RdRp M666 pocket is lined by 24 amino acid sidechains (figure 3, Zenodo report).
We then conduct a broad survey of viral proteins from Alpha- and Betacoronaviruses to identify the most conserved druggable sites. We are integrating variability and druggability data to infer the most promising strategies for developing broad-spectrum viral inhibitors. This analysis can be valuable in the context of the emergence of future coronaviruses circulating in bat reservoir species.
To assess this site’s genetic variability, we first looked at 27 reviewed sequences from Uniprot, of which 6 are from the Alphacoronavirus genus and 21 from the Betacoronavirus genus. Altogether, eight out of twenty-four residues at M666 were identical across the 27 entries, equivalent to 33% identity at this site (figure 4, Zenodo report).
As a follow-up to assessing the genetic variability among Alphacoronavirus and Betacoronavirus entries, our collaborator, Nicola De Maio, from Nick Goldman’s lab at the European Bioinformatics Institute has identified the non-synonymous mutations at M666 sites across more than 15000 SARS-CoV-2 samples. Altogether he identified non-synonymous mutations at 10 unique amino acid residues consisting of 13 unique non-synonymous variants. In table 1 on our Zenodo report, there are details about the mutation events underlying these variants.
In terms of druggability, the M666 pocket (Dscore=1.028) is superior to the RdRp active site (where remdesivir binds) (Dscore=0.859). While the active site is highly conserved (83% identity), the M666 pocket is poorly conserved (33% identity). Similarly, across SARS-CoV-2 samples at the M666 site, there is high genetic variability compared to the remdesivir site, which is more genetically stable; only 6 out of 48 residues were mutated.
Remdesivir use for COVID-19 is only recommended for hospitalized patients who need supplemental oxygen therapy (Clinical Trials.gov number, NCT04280705), and only in those severe cases, remdesivir was superior to placebo in shortening the time to recovery. (Goldman et al., 2020) This phase3 study showed that despite remdesivir, there was still high mortality because of COVID-19. Therefore, clinically we still need a better drug to treat COVID-19 through different approaches. (https://www.nejm.org/doi/full/10.1056/NEJMoa2007764)
Targeting the M666 provides an alternate strategy to target RdRp function by disturbing its interaction with a key accessory factor, nsp8. It is important to note the high genetic variability of this site among coronaviruses and SARS-CoV-2 samples, which deprioritizes it as a target for pan-coronavirus anti-SARS-CoV-2 drugs.
You can view this report on Zenodo as well.
Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project.
References:
Yin, W., Mao, C., Luan, X., Shen, D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y., Tian, G., Jiang, H., Tao, S., Shen, J., Jiang, Y., Jiang, H., Xu, Y., & Xu, H. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science, 368(6498), 1499-1504. https://doi.org/10.1126/science.abc1560
Goldman, J. D., Lye, D. C.B., Hui, D. S., Marks, K. M., Bruno, R., Montejano, R., Spinner, C. D., Galli, M., Ahn, M., Nahass, R. G., Chen, Y., SenGupta, D., Hyland, R. H., Osinusi, A. O., Cao, H., Blair, C., Wei, X., Gaggar, A., Brainard, D. M., & Subramanian, A. (2020). Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. The New England Journal of Medicine. https://doi.org/10.1056/NEJMoa2015301
