I’ve been getting my stocks in order recently and catching up on some other work which has been both busy but not the most thrilling in terms of experimental results. As such I’ve not had much time to write up a blog post so I’ll try and get back on top of that although it’s not likely to be that world shattering.
I’m still trying to purify versions of ALK2 where various phosphorylation sites at the top of the molecule have been mutated out so that we can see what influence each of the phospho-sites has on the overall activity of ALK2 and hopefully from that work out which sites are essential for ALK2’s function and which are merely ‘nice to haves’ or even not necessary at all. This will hopefully then combine with our study of the mutant version of ALK2 to try and see whether the mutation that causes FOP is interfering with this phosphorylation activation. Anyway – that’s the ultimate goal, in the mean time I’ve been slowly purifying more constructs so that I can do a big experiment comparing all the different constructs at the same time. The main reason I want to do them all at once is to try and minimise the variability that can occur between experimental runs despite best efforts to make everything identical. If I’m actually using the same stock of partner proteins and ATP and metal ions and incubating them for exactly the same time it makes the comparisons between the mutants easier and less likely to contain artefact errors.
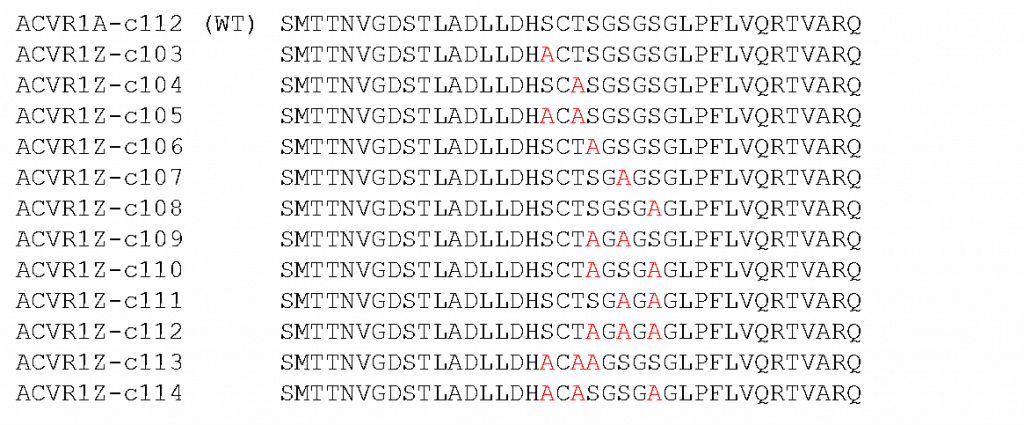
Figure 1: The different ACVR1 (ALK2) GS loop phosphomutants generated and their associated construct id’s. The top row shows the sequence of the wild type protein. Mutations are shown in red.
So – with that I’ve been stockpiling the various mutants as they come through the expression pipeline at the SGC. This time around I’ve been purifying three more constructs – ACVR1-105, 106 and 109 (constructs 107 and 108 and beyond are still to be sorted). (Just a quick reminder that ALK2 and ACVR1 are different ways of referring to the same thing, technically ALK2 is the protein and ACVR1 is the gene)
I had a bit of a problem with some of them – I’m not sure why but they seem to require more Nickle resin than usual to purify compared to the wild type construct – I’m not sure if that’s because the mutation makes it harder for the tag to contact the resin or if it is because there is more background contamination than usual in this particular line of cells or whether it’s something else. Luckily they can be rescued through re-incubating the sample with more nickle to pull out the protein and continuing on as usual. This is, while annoying, not a complete loss so while I’m keen to find out why it’s behaving differently it’s not ruined my purification prep.
There are still a couple of constructs left to purify before I run the big experiment and there are a few other small optimisations I need to do as well but I’m getting closer to really getting to the bottom of this question.
If you want to read more about the exact purification process then please have a look over on Zenodo.
