This is a summary of a detailed analysis that is available here.
Helicases are motor proteins that separate double helices through the hydrolysis of ATP analogues (Fairman-Williams et al., Current Opinion in Structural Biology, 2010). They constitute one of the largest enzyme groups, as multiple varieties of helicases are present in all forms of cellular life (Chamieh et al., Gene, 2016). Helicase activity is required for RNA and DNA metabolism, consequently, malfunction or dysregulation within the enzyme can induce a variety of diseases such as genomic syndromes, premature ageing, and cancer (Patel and Donmez, Journal of Biological Chemistry, 2006).
Since, helicases play a critical role in many biological processes within virtually all cellular life, they are prime targets for drug discovery and development. The main objective is to discover potent and specific helicase inhibitors that can lead to the development of drugs and therapeutics targeting infectious pathogens or cancer cells.
I have decided to analyze the helicase domain, the only domain present in all helicases, as a potential target for drug discovery (Young and Karbstein, Methods in Enzymology, 2012). However, the binding pockets and the structural diversity within this domain has not been analyzed. Therefore, as a first step, I begin with creating a multiple sequence alignment to determine structural similarities and differences between human and four viral families with pandemic potential. From this multiple sequence alignment, I constructed a phylogenetic tree to gain insights into the function, structure, and evolution of the helicase domain (Figure 1).
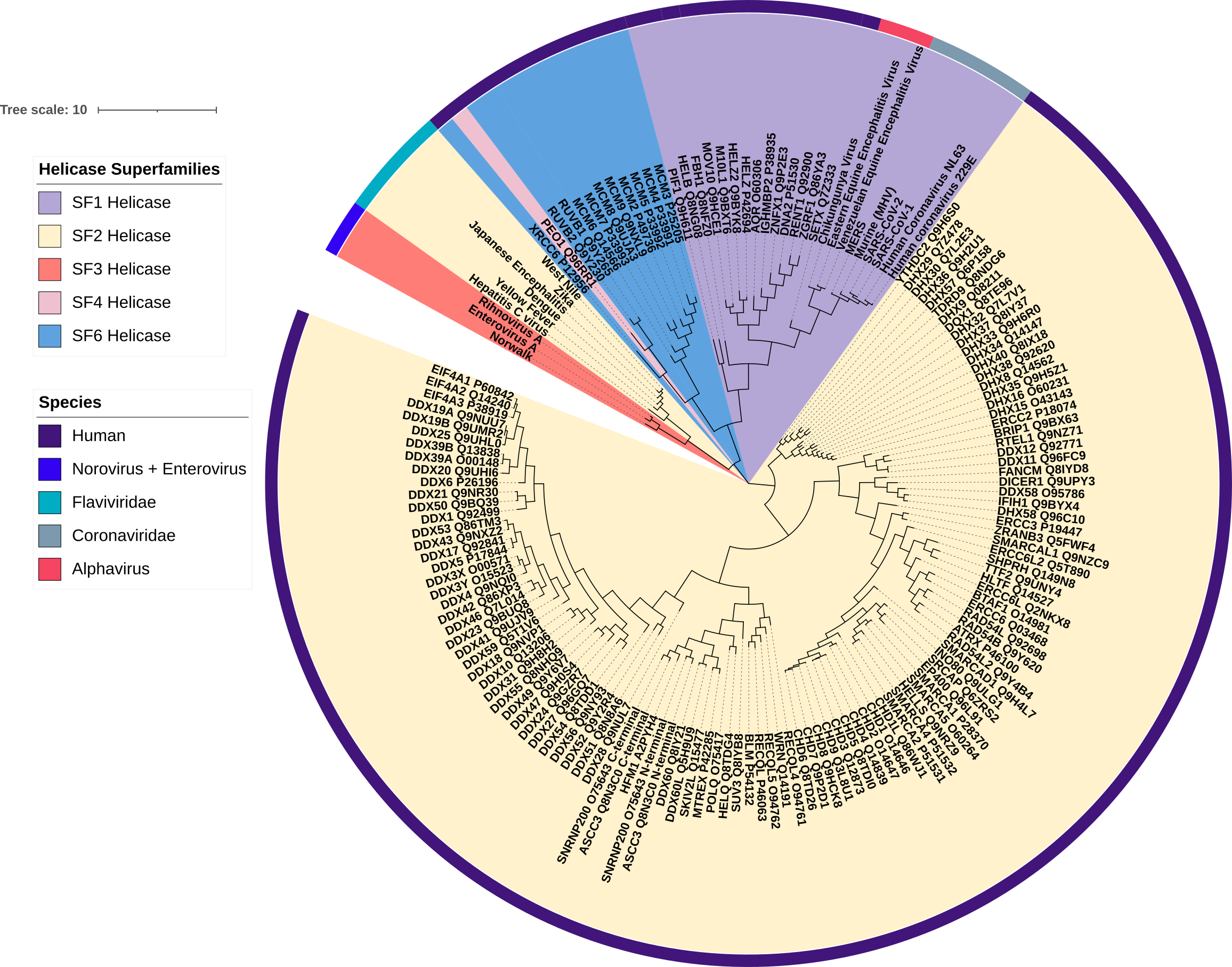 Figure 1. A phylogenetic tree representation of the human and viral helicases. The branches on the phylogenetic tree represent the genetic transmission from one generation to the other. The length of the branches indicate genetic change, i.e. the longer the branch, the more divergence or genetic change has occurred. The different colours highlighting the helicases represent their helicase superfamily, while the colours surrounding the tree indicate which species or viral family the helicase originates from.
Figure 1. A phylogenetic tree representation of the human and viral helicases. The branches on the phylogenetic tree represent the genetic transmission from one generation to the other. The length of the branches indicate genetic change, i.e. the longer the branch, the more divergence or genetic change has occurred. The different colours highlighting the helicases represent their helicase superfamily, while the colours surrounding the tree indicate which species or viral family the helicase originates from.

Beautiful work. From your sequence alignment, it appears that the human Upf1-like SF1 helicases are the closest to the viral SF1 helicases. Certainly much closer than any of the SF2 family helicases. Are there any known small molecule inhibitors of Upf1-like helicases?
Hi Tim,
Unfortunately, there does not seem to be any inhibitors of human Upf1-like helicases.
I agree, this figure is great. Out of curiosity, where would the DNA helicase of the Human Herpes Virus would fit into this tree?
Hi Holli-Joi,
I found that HSV (PDB ID P10189) is quite different from the other SF1 helicases. Here is the tree I’ve created with HSV highlighted: https://drive.google.com/file/d/1A4rAOpgbgGueQhyL4H_0ph-YHoEmYn1P/view?usp=share_link
Interesting. Thanks so much for looking into this!