USP5 is one of many USP proteins that contains a zinc finger ubiquitin binding domain (Zf-UBD). My goal is to develop small molecules that inhibit USP5 selectively. So, it’s important to make sure that the pocket where the compound will bind is sufficiently and structurally different from other USPs. I did a multiple sequence alignment of the USP Zf-UBD sequences against the histone deacetylase 6 (HDAC6) Zf-UBD using ICM-Pro. HDAC6 Zf-UBD has structural similarity to USP Zf-UBD proteins, and because its crystal structure with small molecule inhibitors has been solved (5KH3, 5B8D) I can use it to compare binding pocket residues of the USP Zf-UBDs. More details can be found on Zenodo and the free software (ICM Browser) to view the structural data can be downloaded from Molsoft. HDAC6 Zf-UBD has two adjacent pockets- a primary ubiquitin binding pocket and an adjacent secondary pocket.
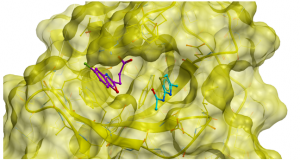
Figure 1. Structures of different ligands binding HDAC6 Zf-UBD at ubiquitin binding pocket (magenta: 5KH3) and adjacent secondary pocket (cyan: 5B8D)
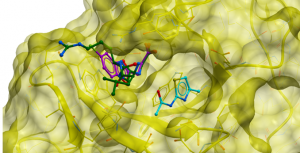
Figure 2. Structure of ubiquitin peptide RLRGG (green: 3GV4) binding in primary binding pocket similarly to ligand (magenta: 5KH3)
The multiple sequence alignment of the USP Zf-UBDs against HDAC6 Zf-UBD shows that the primary binding pocket residues are more conserved than the secondary binding pocket residues. This is encouraging, but we will need to find inhibitors that extend beyond the ubiquitin binding pocket, as was recently done with HDAC6 [1].

Figure 3. Multiple sequence alignment of USP Zf-UBDs against HDAC6 Zf-UBD. Residues highlighted in magenta are more conserved and lining the primary pocket. Residues highlighted in cyan are less conserved and lining the secondary pocket.
[1] Harding J. R., et al. Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin. J Med Chem, 2017. 60(21): 9090-9096
