Following the work on the SARS-CoV-2 main protease, we selected NSP16 as the next target of interest. But what is NSP16?
The coronavirus genome size is large, about 30 kb single-stranded positive RNA. The RNA molecule is 5’-capped and has 3’-poly-A tail. The viral RNA is the molecular template that is translated to make viral proteins. The products of the first open reading frame are polyprotein 1a and 1ab which are further processed to make the non-structural proteins (NSPs). The assembly of the NSPs forms the replication-translation complex (RTC), and among those NSPs, there are two enzymes involved in viral mRNA capping. The capping of the viral mRNA is a critical viral defense mechanism against the immune system. One of these two enzymes is NSP16 which is a SAM-dependent m7GpppA-specific 2’-O-methyltransferase (2’-O-MTase) which means that it uses the cofactor, S-adenosyl-L-methionine (SAM) as the methyl donor. (1) NSP16 forms a stable dimer with NSP10, then NSP10-NSP16 mediates 2’-O-methylation of coronavirus mRNA to prevent viral recognition by the host to avoid immune response during mRNA translation. (1, 2) This viral mechanism makes NSP16 protein an appealing target for anti-SARS-CoV-2 therapies.
In this post, I will briefly walk you through the steps for finding the active site of this protein and its druggability measure. Then, I will explain how we picked the surrounding residues that line the NSP16 catalytic pocket.
I use NSP16 in this post to refer to SARS-CoV-2 SAM-dependent m7GpppA-specific 2’-O-methyltransferase.
The very first step was to identify the potential drug-binding sites on NSP16 (PDB: 6wvn). We used ICM PocketFinder (Molsoft, SD) to identify the pockets. The druggability analysis was done using SiteMap (Schrodinger, NY). The results are shown in Figure 1. The druggability score (Dscore) of the catalytic site of NSP16 is 0.98 which is indicative of a druggable site.
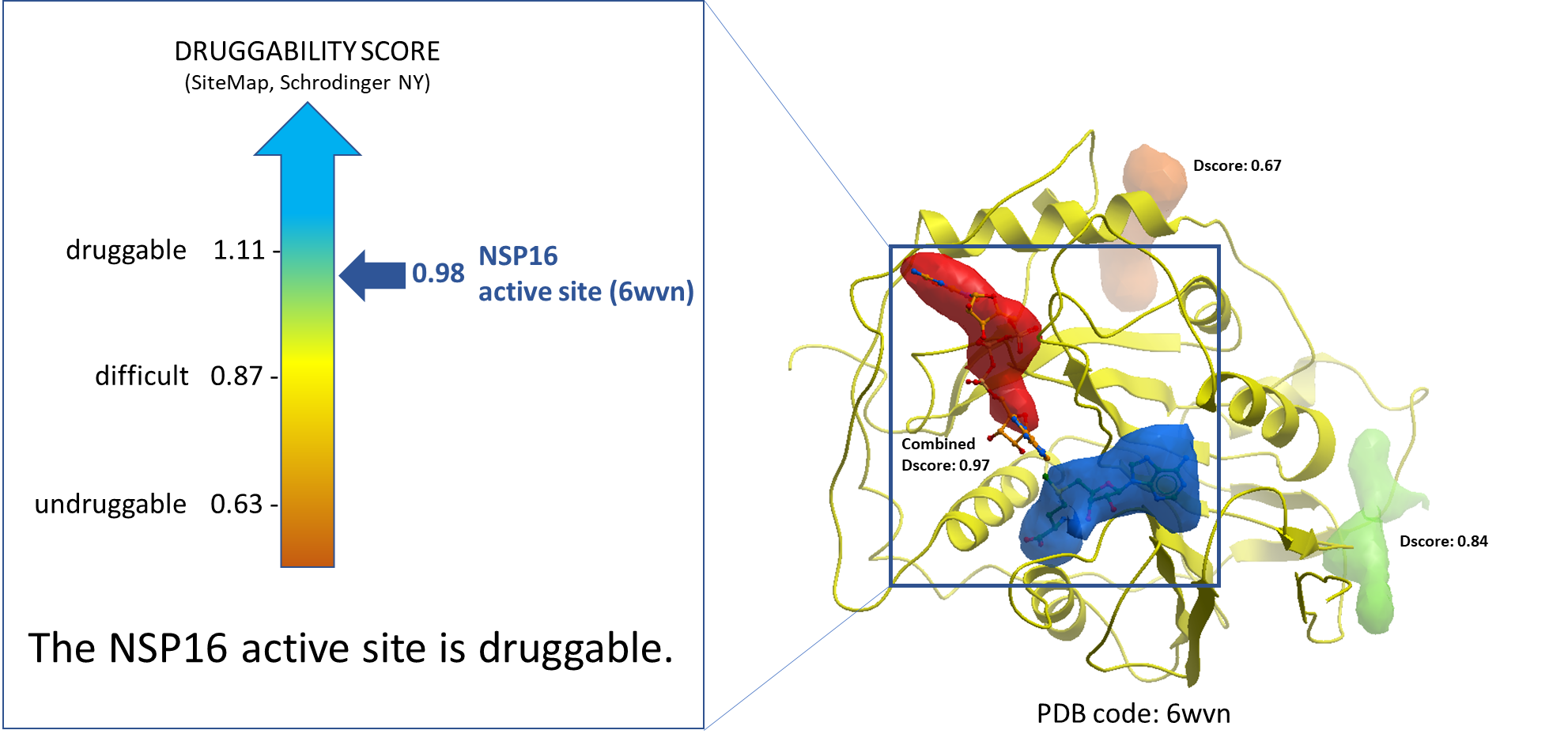
Figure 1. Druggability analysis of NSP16 catalytic site. The blue and the red pockets show the catalytic site of the NSP16 molecule, where the blue pocket is occupied by SAM, and the red pocket is occupied by a substrate RNA molecule.
Next, to determine the residues that line the catalytic site, we picked the sidechains of the protein facing towards the pocket within its 2.8 Å vicinity. At the end, we selected 39 residues that met these criteria. In figure 2 those residues are shown in sticks representation. Here is a full list of these 39 residues: K6822,C6823,D6824,L6825,Y6828,N6841,K6844,Y6845,H6867,F6868,G6869,A6870,G6871,S6872,P6878,G6879,S6896,D6897,L6898,N6899,D6912,C6913,D6928,M6929,Y6930,D6931,P6932,T6934,K6935,V6937,F6947,K6968,T6970,E6971,H6972,N6996,S6999,S7000,E7001.
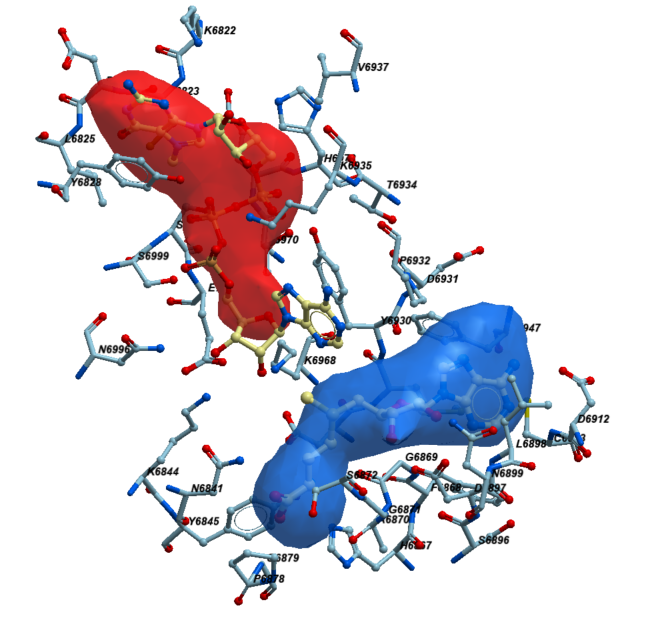
Figure 2. The neighboring residues lining the catalytic pocket of NSP16.
To view the detailed steps, please see my Zenodo report here.
In my next post, I will show the sequence diversity across UniProt entries from the Alpha- and Betacoronavirus genera and map that to the NSP16’s catalytic site using its crystal structure.
As always, I would be happy to hear your comments. Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project.
References:
- Rosas-Lemus, M. et al. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv 2020.04.17.047498; DOI: https://doi.org/10.1101/2020.04.17.047498. Apr 2020.
- Hyde, J. L., and Diamond, M. S. (2015) Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology 479-480, 66–74. DOI: https://doi.org/10.1016/j.virol.2015.01.019
