Previously we reported our analysis on the structural diversity of binding pockets found on the SARS-CoV-2 Main protease, methyltransferase, and papain-like protease. We then shifted our focus on the SARS-CoV-2 macrodomain (nsp3-Mac1).
There are six macrodomain classes based on structural similarity. Most viral macrodomains fall into the MacroD-like family the same as human homologs MacroD1 and MaroD2, which remove mono-ADP-ribosylation (MAR). (Xu et al., 2008)
Non-structural protein 3 (nsp3) is a large multidomain protein encoded by SARS-CoV-2 RNA. Within nsp3, there are three tandem macrodomains, including Mac1, Mac2, and Mac3. In contrast to Mac2 and Mac3, Mac1 is conserved among coronaviruses (CoVs), and it binds to mono- and poly-ADP-ribose and hydrolyzes MAR from target proteins. Mac1 is known to interfere with the immune response, possibly due to its ability to remove ADP-ribose from ADP-ribosylated proteins and RNA, which is part of the host’s immune response’s intricacies. (Alhammad et al., 2020)
PARPs (poly ADP-ribose polymerases) add MAR subunits to their substrates after being induced by interferon to inhibit viral replication. Therefore Mac1’s function is opposite to PARPs since, by de-MARylating target proteins, it contributes to viral evasion from the host defense mechanism (Alhammad et al., 2020).
In a previous study, mutated macrodomain replicated poorly in macrophages in the bone marrow, the primary cells in the innate immune response. This finding shows that macrodomain’s inhibition may help reduce viral replication. (Grunewald et al., 2019)
There is further experimental evidence that supports the role of conserved macrodomain in CoV’s virulence and suppression of immune response. Eriksson and her colleagues showed that mutating a conserved asparagine residue of Mac1 to an alanine residue attenuated the hydrolase activity of Mac1 in SARS-CoV (Eriksson et al., 2008). Additionally, Fehr and his colleagues showed that the asparagine-to-alanine substitution in the hepatotropic mouse hepatitis virus strain A59 (MHV-A59) macrodomain resulted in attenuation in the wild type mice. (Fehr et al., 2014) Furthermore, the asparagine-to-alanine Mac1 mutant also attenuates SARS-CoV viral load in a mouse model of severe respiratory disease. (Fehr et al., 2016) Therefore the viral macrodomain counters host antiviral ADP-ribosylation.
Mac1 is a critical pathogenicity agent in animal models of CoV infection, making it an important virulence factor. In summary, Mac1 could be an attractive target for small-molecule antivirals with the caveat that a highly selective compound can be found to discriminate between the human and the viral macrodomains. (Alhammad et al., 2020)
In this post, I will explain how we identified the ADPr-binding site of Mac1 and its druggability measure, and list the residues lining that site.
Using ICM function PocketFinder (Molsoft, SD) we found the potential drug-binding sites using the nsp3-Mac1 x-ray crystal structure (PDB: 6w02). Then using SiteMap (Schrodinger, NY) we assessed the druggability of the binding sites using the druggability score (Dscore). The ADPr-binding site of nsp3-Mac1 has a Dscore of 0.95 indicative of a druggable site.
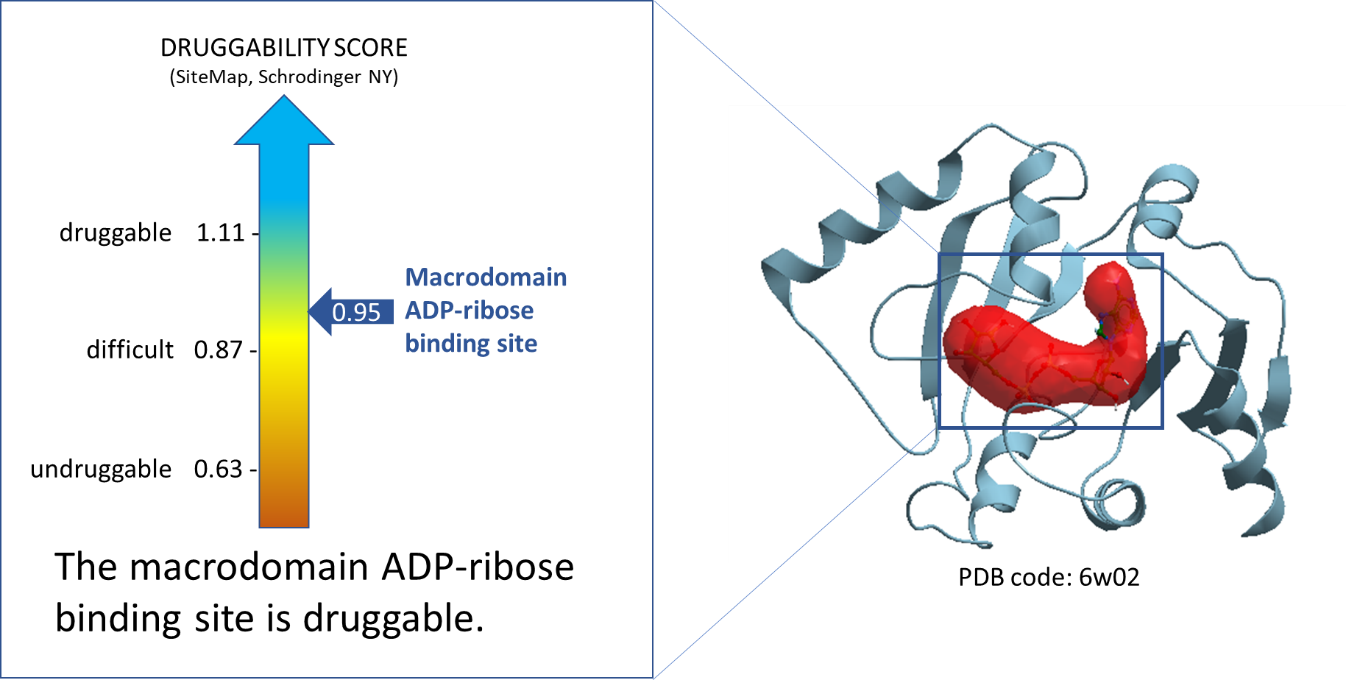
Figure 1. Druggability analysis of SARS-CoV-2 NSP3-Mac1 ADP-ribose binding site. The red pocket is occupied by ADPr.
Using ICM, we then found the amino acid sidechains that face the ADPr-binding pocket within its 2.8 Å vicinity. We identified twenty-nine sidechains in total which include: Asp22, Ile23, Asn37, Ala38, Ala39, Asn40, Hie45, Gly46, Gly47, Gly48, Val49, Ala50, Gly51, Ala52, Gly97, Pro12, Leu12, Leu12, Ser12, Ala12, Gly13, Ile13, Phe13, Pro13, Ala15, Val15, Phe15, Asp15, Leu16. Figure 2 shows these residues in sticks representation around the ADPr-binding pocket.
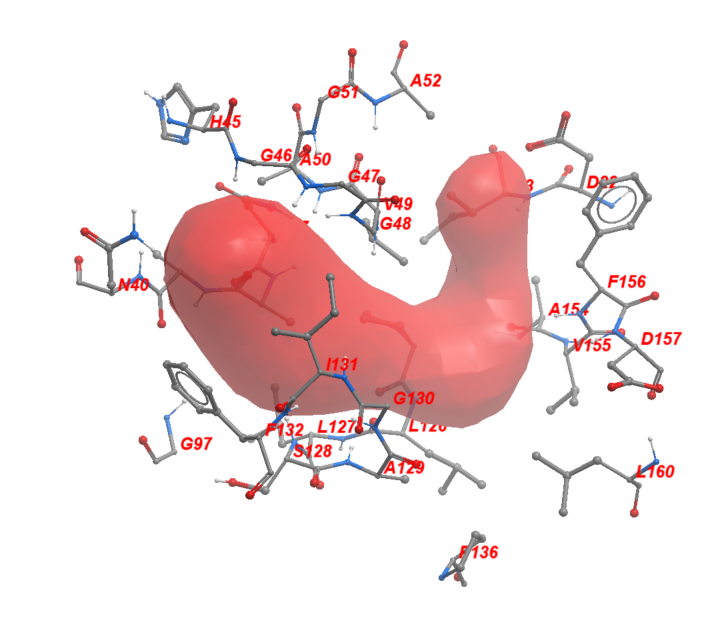
Figure 2. The neighboring residues lining the ADPr-binding pocket of Mac1.
In my next post, I will show the sequence diversity across UniProt entries from the Alpha- and Betacoronavirus genera and map that to the nsp3-Mac1 ADPr-binding site using its crystal structure.
As always, I would be happy to hear your comments. Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for my next post!
References:
Xu, Y., Cong, L., Chen, C., Wei, L., Zhao, Q., Xu, X., Ma, Y., Bartlam, M., & Rao, Z. (2008). Crystal structures of two coronavirus ADP-ribose-1″-Monophosphatases and their complexes with ADP-ribose: A systematic structural analysis of the viral ADRP domain. Journal of Virology, 83(2), 1083-1092. https://doi.org/10.1128/jvi.01862-08
Alhammad, Y. M., Kashipathy, M. M., Roy, A., Gagné, J., Nonfoux, L., McDonald, P., Gao, P., Battaile, K. P., Johnson, D. K., Poirier, G. G., Lovell, S., & Fehr, A. R. (2020). The SARS-Cov-2 conserved macrodomain is a highly efficient ADP-ribosylhydrolase. bioRxiv. https://doi.org/10.1101/2020.05.11.089375
Grunewald, M. E., Chen, Y., Kuny, C., Maejima, T., Lease, R., Ferraris, D., Aikawa, M., Sullivan, C. S., Perlman, S., & Fehr, A. R. (2019). The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLOS Pathogens, 15(5), e1007756. https://doi.org/10.1371/journal.ppat.1007756
Eriksson, K. K., Cervantes-Barragán, L., Ludewig, B., & Thiel, V. (2008). Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-Phosphatase, a viral function conserved in the Alpha-like supergroup. Journal of Virology, 82(24), 12325-12334. https://doi.org/10.1128/jvi.02082-08
Fehr, A. R., Athmer, J., Channappanavar, R., Phillips, J. M., Meyerholz, D. K., & Perlman, S. (2014). The nsp3 Macrodomain promotes virulence in mice with coronavirus-induced encephalitis. Journal of Virology, 89(3), 1523-1536. https://doi.org/10.1128/jvi.02596-14
Fehr, A. R., Channappanavar, R., Jankevicius, G., Fett, C., Zhao, J., Athmer, J., Meyerholz, D. K., Ahel, I., & Perlman, S. (2016). The conserved coronavirus Macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio, 7(6). https://doi.org/10.1128/mbio.01721-16
