Previously, I provided a brief overview on SCN1A related Dravet Syndrome (DS) and the need for larger gene therapy vectors or alternative methods to deliver functional copies of SCN1A to affected cells in DS. The goal of my project is to explore one of these alternative methods, specifically a split-intein mediated protein trans-splicing method, that could reduce the size of the passenger (SCN1A expression cassettes) for gene therapy vectors. In this post, I will talk about the overall design for this method.
To give you a little more background about inteins: Inteins (also known as intervening proteins) are basically mobile protein elements that are translated and embedded within another protein sequences. You can think of them as single-use, single-turnover enzymes that catalyze their own excision from a flanking precursor “exteins” protein sequences while concurrently joining the remaining exteins protein fragments together through the formation of a new peptide bond (Fig. 1).
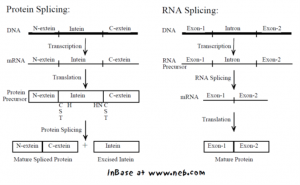
Figure 1. Intein-mediated protein splicing vs. RNA splicing. Image source: Perler, F. B. (2002). InBase, the Intein Database. Nucleic Acids Res. 30, 383-384.
There are two types of inteins – cis splicing inteins that are embedded within protein sequences and catalyze protein splicing in cis and split- or trans-splicing inteins where the enzyme is naturally split and expressed into two parts. These two fragments of split-intein can associate with each other to form a catalytic active enzyme that can catalyze protein splicing in trans. Briefly, my project aims to split the SCN1A expression cassette into two parts and make use of the unique split-intein mediated trans-splicing capability to rejoin the expressed SCN1A fragments together (Fig. 2)
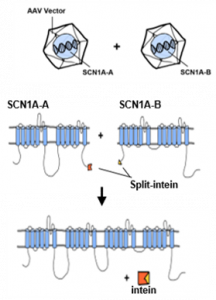
Figure 2. A basic design of split-intein mediated SCN1A protein trans-splicing
So, here goes the basic components and design of this cloning experiment. We are going to express SCN1A as two protein fragments (split-N terminal and split-C terminal SCN1A), each fused with split-N-term and C-term intein sequence (Fig. 2), respectively.
But… where do we split the protein?
One of the factors taken into consideration in designing the split sites is that for efficient protein trans-splicing, the split sites or the sites where split-intein sequences are inserted need to be surface exposed. SCN1A is a multipass membrane proteins with four homologous (unexposed) transmembrane domains (DI to DIV), each domain is connected to the next domain by large cytoplasmic loops (Fig. 3). In fulfilling this condition, we are left with the N and C-terminal cytoplasmic domain and the cytoplasmic loops connecting DI to DII, DII to DIII, and DIII to DIV to work with. Among these cytoplasmic regions, splitting cytoplasmic loops within and between DII to DIII can result in a significant reduction in the size of SCN1A expression cassettes.
Additionally, to initiate efficient splicing reaction, split-inteins require a certain combination of amino acids flanking the intein sequences. For instance, split DnaB intein from Synechocystis needs to be flanked by a glycine at the N-terminus of the intein and a serine at C-terminus of the intein to catalyze protein trans-splicing whereas Vma intein from S. cerevisiae requires a glycine and cysteine amino acids combination. In addition to previous considerations, these specific requirements of amino acids combinations for efficient and seamless protein trans-splicing helped us further narrow down a short list of potential SCN1A split sites.
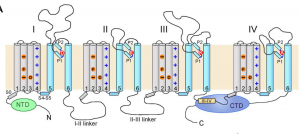
Figure 3. A general topology of eukaryotic voltage-gated sodium channels.
Image source: Shen, Huaizong, et al. “Structure of a Eukaryotic Voltage-Gated Sodium Channel at near-Atomic Resolution.” Science, American Association for the Advancement of Science, 3 Mar. 2017, science.sciencemag.org/content/355/6328/eaal4326.long.
How can we tell whether two split-SCN1A fragments trans-spliced?
To screen and identify constructs that have successfully reconstituted, I will attach split-GFP sequences to the N and C termini of the split-N-terminal-SCN1A intein and split-C-terminal-SCN1A-intein, respectively. Split-GFP is composed of two complementing split fluorescent protein fragments that irreversibly associate with each other to generate a fluorescent signal. When expressing both of these constructs in cells, we predicted that the split-GFP sequence would help bring the two split-proteins together (Fig. 4.) The split-intein would interact with each other to form an active enzyme and catalyze the excision of both itself and GFP protein to the cytoplasm. In addition to using the cytoplasmic GFP signal as a visual positive marker for trans-splicing, the split-GFP system could also be utilized as an alternative way to link two SCN1A fragments together.
So far, I have generated a pair of split-GFP construct to examine one of the split-site. You can find more details of these experiments on zenodo. Going forward, I will be transfecting these constructs into HEK293 cells to examine the following questions: Do these split-protein fragments reconstitute into full-length SCN1A. If so, do they localize and function similar to wild-type SCN1A? Does overproduction of split-protein fragments cause a dominant negative effect on wild-type SCN1A?
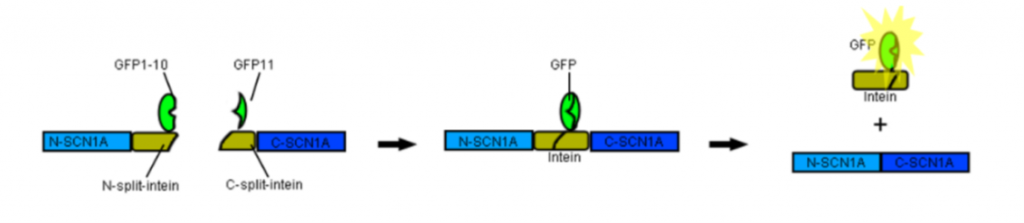
Figure 4. A fluorescence reporter system for identifying SCN1A trans-splicing.
