The experimental set-up and results covered in this post can be found here: 10.5281/zenodo.1495129.
Apologies for the long delay between this post and my last post, it really has been quite some time! As postdocs, we generally have several projects running simultaneously and we have to split our time between these projects. I am no exception and lately I have been caught up in trying to wrap up several projects. Besides my work in HTT, I am involved in screening various cell lines against our SGC probe library and identifying probes that have a differential effect in these cell lines. At the SGC, chemical compounds only qualify as probes if they have an in vitro potency of <100 nM, >30-fold selectivity vs other subfamilies, and have significant on-target cell activity at 1 µM. These probes are essential for the early stages of drug development as positive hits in our screens allow us to identify potential drug targets, protein families, and pathways for which we can further investigate in terms of disease mechanism and therapeutic strategies. These probes identified as positive hits in our screens may then be further developed and refined as drugs which may be used in the clinics eventually. You can read more about the SGC’s probes here: https://www.thesgc.org/chemical-probes.
In one of the other projects I’m working on, we identified PRMT5 as a potential therapeutic target in glioblastoma through the action of GSK591 and LLY283, 2 probes that inhibit PRMT5 through different and independent mechanisms. The inhibition of PRMT5 as a therapeutic strategy is not novel in itself, with several other groups having already published on this.
PRMT5 as a druggable target for glioblastoma therapy.
Banasavadi-Siddegowda YK, Welker AM, An M, Yang X, Zhou W, Shi G, Imitola J, Li C, Hsu S, Wang J, Phelps M, Zhang J, Beattie CE, Baiocchi R, Kaur B.
Neuro Oncol. 2018 May 18;20(6):753-763. doi: 10.1093/neuonc/nox206.
Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma.
Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, Neupane R, Fenoglio S, Cahill DP, Wakimoto H, Agar NYR, Yaffe MB, Sharp PA, Hemann MT, Lees JA.
Cancer Cell. 2017 Oct 9;32(4):411-426.e11. doi: 10.1016/j.ccell.2017.08.018. Epub 2017 Sep 28.
However, we had some interesting findings through the RNA-sequencing done on cells treated with these PRMT5 inhibitors. Serendipitously, HTT emerged as one of the genes whose expression was consistently downregulated with PRMT5 inhibition! I tested these PRMT5 inhibitors in the HeLa cells I used to generate the 3 stable HTT knockdown lines previously (read more about that here: https://openlabnotebooks.org/generating-htt-knockdown-models/) and looked at how the HTT protein levels were affected in these lines.
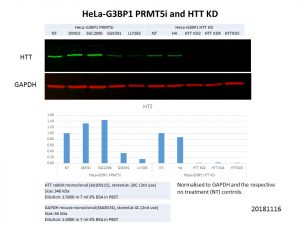
HTT protein levels were clearly decreased with PRMT5 inhibition! I ran these samples alongside the stable HTT knockdown lines and although the HTT levels did not decrease as drastically as with the knockdowns, the decrease with the PRMT5 inhibitors is still substantial. I also included SGC2096, the inactive control to GSK591, and HTT protein levels were not affected by it, showing that it is indeed through the activity of PRMT5 that HTT levels were decreased.
The mechanism by which HTT is downregulated by PRMT5 inhibitors is still unclear and it will be very interesting to further investigate the link between HTT and PRMT5. As of now, the only publication I can find linking HTT to PRMT5 is the following:
PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD).
Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA.
Cell Cycle. 2015;14(11):1716-29. doi: 10.1080/15384101.2015.1033595.
This paper showed the colocalisation and interaction of HTT and PRMT5 and shows that this interaction results in the stimulation of PRMT5 activity by HTT in vitro. Mutant HTT has a greater affinity for PRMT5; however, it has a reduced ability to stimulate PRMT5 activity, as demonstrated by the reduction of symmetric dimethylation of arginine (SDMA), which is mainly mediated by PRMT5, of histones H2 and H4. Decreased levels of SDMA on known PRMT5 targets were detected in HD brain samples, suggesting that the reduction in PRMT5 activity may contribute to the pathogenesis of HD. The knockdown of JMJD6, which removes arginine demethylation, was able to rescue the phenotype of increased cell death seen with mutant HTT, further suggesting that the restoration of SDMA to normal levels may be a potential therapeutic strategy.
However, our findings that PRMT5 inhibition itself can affect the expression of HTT at the protein level suggests that there might be a feedback loop in play between PRMT5 and HTT. It will be interesting to further investigate how the levels of SDMA and the other proteins involved are affected with PRMT5 inhibition and elucidate the mechanisms involved in this process. It should also be noted that our results only reflect the expression of wild-type HTT and not mutant (expanded) HTT, and it will be highly interesting and of great clinical importance to find out if PRMT5 inhibition is able to decrease the protein levels of mutant HTT similarly to how it affects wild-type HTT.

Very cool, Jolene. What PRMT5 inhibitor concentration did you use?
… And looking forward to seeing the effect on mutant HTT levels!
Thanks Matthieu! I used both GSK591 and LLY283 at 1 µM, which was the concentration we screened the compounds at. However, by looking at the reduction in HTT protein levels, LLY283 seems to be more potent than GSK591 and was able to induce a greater downregulation of HTT protein levels at the same concentration asGSK591. This might be due to the difference in the way GSK591 and LLY283 work as they inhibit PRMT5 through different mechanisms, or it might simply be due to differences in their chemical properties, which affect how easily they can enter the cells.
Are you planning to test the inhibition of HTT expression in neurons? I would be curious to see what it does as I think it’s involved in neural stem cell self renewal…
We tried the HTT shRNAs in GBM stem cells but didn’t observe any phenotypes. In contrast, PRMT5 inhibition shows a strong effect on GBM stem cell self-renewal.