This week one of the things I’ve been working on is the final set of mutations in the GS domain of Alk2 to help us unpick the influence of phosphorylation on function.
That, however, is a bit of a mouthful so let’s unpack it a bit.
I said in my last blog post that we know that for Alk2 to phosphorylate SMAD1 it must contain the GS domain. This is a short glycine and serine rich loop of the protein that comes before the main kinase domain. The serine residues in this region can be phosphorylated and this seems to be key to activating Alk2. The pattern of phosphorylation of these residues may well be important and split in specific ways between auto phosphorylation and trans phosphorylation by a type II receptor.
In order to work out which phosphorylation’s are key for SMAD activation, we designed mutants that would change the serine residues (where phosphorylation occurs) to alanine residues, thus preventing phosphorylation on that residue. In different combinations this allows us to test which phosphorylations are key to activation.
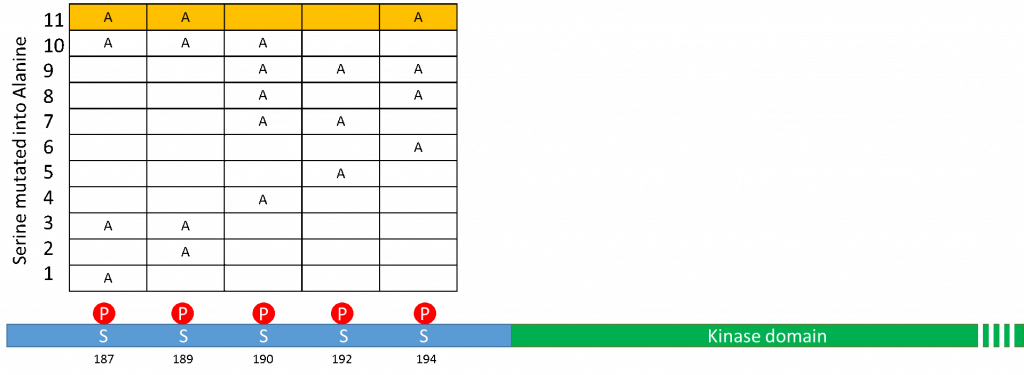
Mutations in the GS domain (blue) of Alk2, immediately prior to the kinase domain (green). The table shows the 11 different mutants that probe the different combinations of phosphorylation events necessary for SMAD1 activation. Row 11 in yellow is the mutant recently cloned.
My colleague George Kerr designed and cloned most of the constructs but moved onto a new role before the final one was made. I’ve now tried to make the final clone (highlighted in yellow) based on her primer designs.
I got colonies from my cloning and picked four to send for sequencing. Sequencing revealed that all four colonies contained the correct mutation and no additional mutations.
These mutant proteins can now be used to do in-cell assays, used as templates to make constructs suitable for expression and from there used in many of the in-vitro experiments I’ve already described to help work out the influence each of these phosphorylation events has.
For more details on the experimental methods please look at Zenodo here.
