One goal of the CaMKK2 project is to explore the chemistry and biology of novel hinge binders. Various hinge binders showing CaMKK2 inhibition have been identified in the literature and through testing of compounds synthesized in the SGC. Of note in this search was a recent publication by Price et al. (Bioorg. Med. Chem. Lett., 2018, 28, 1958–1963), in which molecules based on 7-azaindole (1,2) and 2-amino-pyrimidine (3) (Figure 1) cores showed excellent activity in an enzymatic CaMKK2 assay.
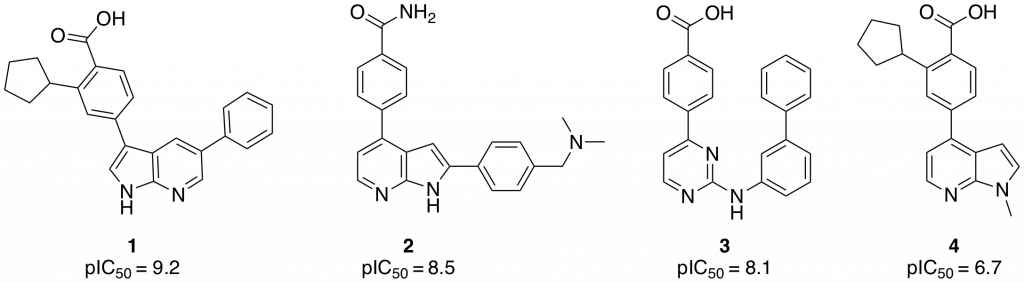
Figure 1: Azaindole based CaMKK2 inhibitors
Although the compounds have good potency, they are promiscuous, showing poor selectivity in a small screen against 48-86 different kinases. Indeed between 15-30% of the kinases in the panel were inhibited with an IC50 <100 nM.
Small modifications to the hinge binding region of pharmacophore can have drastic effects on the binding mode, and therefore the selectivity and potency, of inhibitors. It was decided to probe if N-methylation of the azaindole core could be tolerated in CaMKK2. N-methyl azaindole 4 (Figure 1) was synthesized and sent for testing in a CaMKK2 enzyme inhibition assay. The compounds showed moderate activity giving an IC50 of 183 nM. The compound also gave had good solubility (125 µM) and was membrane permeable. It was decided to synthesis a small library of compounds based on 4 to see if more potent compounds based on this scaffold.
The compounds were relatively easy to access synthetically. N-methylation of 2-iodo-4-chloro-7-azaindole followed by two Suzuki couplings and an ester hydrolysis allowed quick access to the desired compounds. Conveniently, both Suzuki coupling could be carried using microwave conditions with reactions going to completion in 30 minutes (Scheme 1).
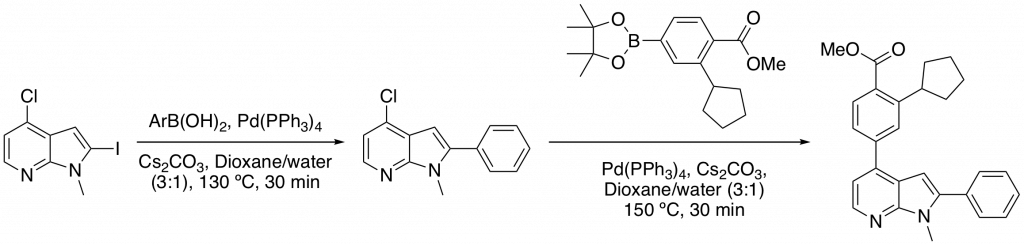
Scheme 1: Sequential Suzuki reactions utilizing a microwave reactor to greatly reduce reaction times.
In choosing what analogues to make in the library I wanted to probe the effects of electron withdrawing and donating groups, lipophilicity and to determine the effect of extending the molecules with morpholino- groups pointing out of the binding pocket towards the solvent. The library of compounds synthesized is shown in Figure 2.
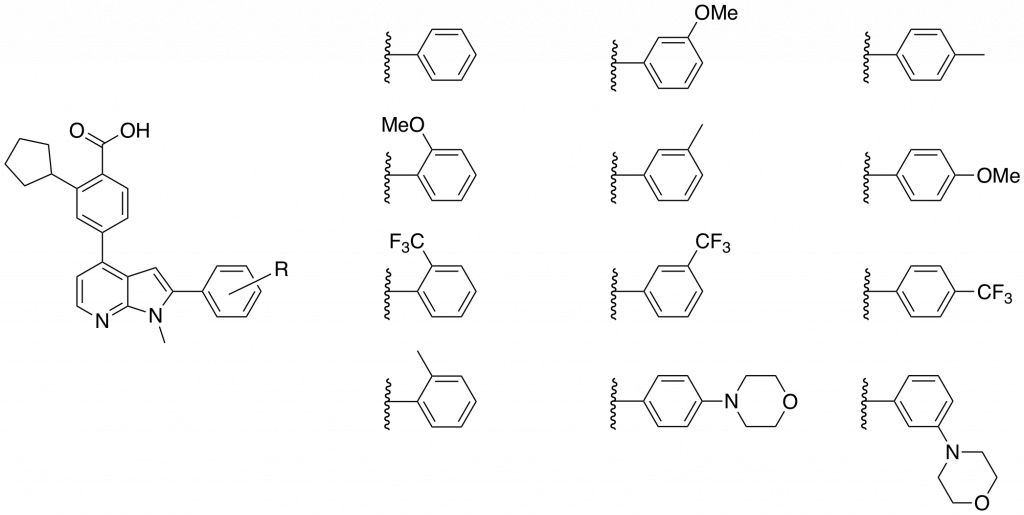
Figure 2: Library of N-methyl-azaindoles synthesized and sent for a CaMKK2 enzyme inhibition assay.
The compounds have been shipped and we are awaiting the results from the enzyme inhibition assay. If any of the compounds have good activity, the best will be sent for a selectivity profiling using KINOMEscan screening platform provided by DiscoverX. From this we will learn if N-methylation can indeed modulate selectivity across the kinome.
