In my last post, I mentioned I would be doing an internship at a biotech company in Montreal, Molecular Forecaster Inc as a part of a NSERC ChemNet CREATE grant. I’ve been in Montreal for about three weeks now and have had a chance to play around with the Forecaster software. I did preliminary docking validation studies for USP5 Zf-UBD using the Forecaster FITTED program. You can find experimental details on Zenodo.
First, I did self-docking studies with USP5 Zf-UBD co-crystal structures to assess the success of ligand docking. Self-docking is a method where a ligand is docked into the protein structure it was extracted from. To evaluate the correctness of the predicted pose, root mean square deviation values (RMSD) are evaluated; the lower the RMSD, the better the docked pose. The predicted binding poses of self-docked ligands was successful (RMSD <2 Å), as shown in Figure 1.
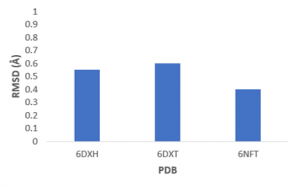
Figure 1. USP5 Zf-UBD self-docking RMSD
An interesting aspect of the Forecaster platform is MATCHUP, where you can dock ligands to a protein and simulate the flexibility of the protein by providing several conformations of structures. The predicted binding poses of ligands docked using MATCHUP was also successful (RMSD <2 Å), as shown in Figure 2, however the docked poses of compound DAT201 and compound DAT180 had higher RMSDs (RMSD >1.5 Å) with flexible docking than rigid self-docking (Figure 3).
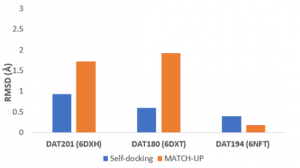
Figure 2. Self-docking (rigid) vs. flexible docking (MATCHUP)
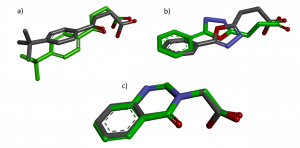
Figure 3. Flexible docked pose (green) versus co-crystallized ligand (grey) a) DAT201 b) DAT180 c) DAT194
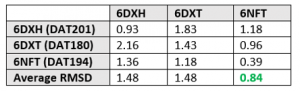
I also performed rigid cross-docking of the available USP5 Zf-UBD structures. Cross-docking is when different ligands are docked to a protein structure from a complex; this is useful when deciding which structure to use for virtual screening. Table 1 summarizes the results of the cross-docking study, where structure 6NFT had the best overall average RMSD.
Table 1. Cross-docking: RMSDs of ligand pose with best energy complex from 3 poses
Lastly, I did a virtual screen of a library of USP5 Zf-UBD compounds that have already been experimentally tested to determine the best ranking docking method (Figure 3). Flexible docking with MATCHUP results in the best overall ranking of USP5 Zf-UBD compounds, when comparing ligand activity and docking scores, as shown by the area under the curve (AUC) values. The AUC of the docking methods are marginally better than random (Figure 3-red (AUC=0.5)) which may be because all compounds in the library tested, include a carboxylic chain that drives binding in the pocket, making it challenging to distinguish between active and inactive ligands. Next, I’ll be using Forecaster’s flexible protein docking with MATCHUP to screen larger commercial libraries in the search for different ligand scaffolds against USP5 Zf-UBD. Stay tuned!
![]()
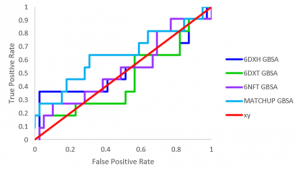
Figure 3. ROC of virtual screen of USP5 Zf-UBD compounds
References
Corbeil C.R., Englebienne P., Moitessier N. Docking ligands into flexible and solvated macromolecules. 1. Development and validation of FITTED 1.0 J. Chem. Inf. Model. (2007), 47, 2, 435-449
Corbeil C.R., Moitessier N. Docking ligands into flexible and solvated macromolecules. 3. Impact of input ligand conformation, protein flexibility, and water molecules on the accuracy of docking programs J. Chem. Inf. Model. (2009), 49, 4, 997-1009
Moitessier N., Pottel J., Therrien E., Englebienne P., Liu Z., Tomberg A., Corbeil C.R. Medicinal Chemistry Projects Requiring Imaginative Structure-Based Drug Design Methods Acc. Chem. Res. (2016), 49 (9), 1646-1657
