Hi all, today’s post covers some concepts that I have previously described: expression and purification of protein constructs, and testing ligands in a displacement assay.
If you’ll recall, I tried to develop and optimize a fluorescence polarization (FP) displacement assay for the ZnF-UBD of USP5 for some time but found that FP was a no go to screen libraries due to a low dynamic range. For the past couple of weeks, I’ve been working on purifying biotinylated USP5 protein constructs (full-length and ZnF-UBD) to use them in a different displacement assay. You can see details of these experiments on Zenodo.
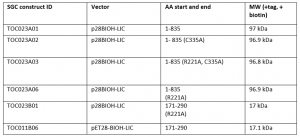
I was successful in purifying various USP5 constructs with a biotin tag. I expressed and purified the constructs summarized in Table 1. Constructs 1-835 are the full length protein, while constructs 171-290 are the ZnF-UBD that I have previously crystallized in complex with my screening hits. R221A mutants were used as controls for the displacement assay. R221 is an important residue in the binding pocket of the ZnF-UBD that co-ordinates ubiquitin binding. A mutation of the arginine (R) to an alanine (A) should prevent binding of ubiquitin (and of my inhibitors) to the ZnF-UBD1,2. C335A is a mutation of the catalytic cysteine residue that carries out the cleavage of polyubiquitin chains3.
Table 1. USP5 construct Summary
The workflow of the displacement assay is summarized in Figure 1. A biotinylated protein is captured to a streptavidin coated plate. Excess protein is removed through washes and a fluorescently labeled ubiquitin (FITC-UBQ) is displaced by a competing ligand resulting in a decrease in fluorescence after wash steps to remove unbound ligands.
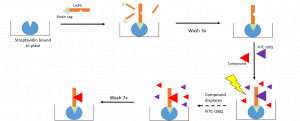
Figure 1. Workflow of Displacement Assay
I tested two ligands) in the displacement assay: unlabelled UBQ, and cpd 21 (Figure 2).
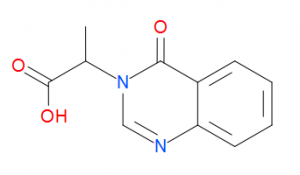
Figure 2. Compound structure of cpd 21
Both UBQ and cpd 21 displaced FITC-UBQ from the ZnF-UBD from FL USP5 (Figure 3). A mutation of the R221 residue to an alanine resulted in no capture of FITC-UBQ and thus no displacement for UBQ and cpd 21 for both the ZnF-UBD and FL USP5.
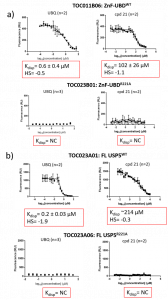
Figure 3. Displacement Assay summary of a) USP5 ZnF-UBD: TOC011B06 (WT-wild type) and TOC023B06 (R221A) and b) FL USP5: TOC023A01 (WT) and TOC023A06 (R221A). HS= hill slope, NC= not calculated.
It should be noted these are preliminary results, and the displacement curves are still quite noisy. Sources of error could include variation of the levels of streptavidin in each well of the 384-Greiner plate. For example, there may be more protein and FITC-UBQ captured in one well over another. This is why it will be important to repeat the experiment several times before having confidence in the reported Kdisp values.
Nonetheless, this is an important finding for the USP5 project. ZnF-UBD ligands are able to displace ubiquitin in both the ZnF-UBD and in the full length protein. Next, I’ll be repeating the experiment several more times to tighten up the data.
References
(1) Reyes-Turcu, F. E.; Ventii, K. H.; Wilkinson, K. D. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 2009. https://doi.org/10.1146/annurev.biochem.78.082307.091526.
(2) Reyes-Turcu, F. E.; Horton, J. R.; Mullally, J. E.; Heroux, A.; Cheng, X.; Wilkinson, K. D. The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin. Cell 2006. https://doi.org/10.1016/j.cell.2006.02.038.
(3) Scott, D.; Layfield, R.; Oldham, N. J. Structural Insights into Interactions between Ubiquitin Specific Protease 5 and Its Polyubiquitin Substrates by Mass Spectrometry and Ion Mobility Spectrometry. Protein Sci. 2015. https://doi.org/10.1002/pro.2692.
