In my previous post I mentioned that there is controversy if HDAC11 can deacetylate histones. I decided to look at the effect of overexpression of HDAC11 on exogenous histone acetylation in cells. In my preliminary results I noticed that exogenous GFP-tagged histones, one day post transfection, have very low levels of acetylation; therefore I decided to add to the system some histone acetyltransferases (p300, GCN5, MYST1, PCAF). My results indicate that overexpression of HDAC11 decrease basal acetylation levels of exogenous histones as well as decrease the acetylation increase caused by histone acetyltransferases. Since we do not know if the observed effect is caused by catalytic activity of HDAC11, I generated catalytic HDAC11 mutants Y304H and H142,143A (previously published PMID:30819897, 28927261), which will be used for validation.
EXPERIMENTAL DESIGN:
HEK293T cells were co-transfected for 20 h in 12 well format with 0.1 µg histone-GFP and:
1.empty vector (0.9 µg)
2.empty vector (0.1 µg) + Hdac11-FLAG (0.8 µg)
3.HAT-FLAG (0.1 µg) + empty vector (0.8 µg)
4.HAT-FLAG (0.1 µg) + Hdac11-FLAG (0.8 µg)
Histones: H3, H4, H2A, H2B
HATs: MYST1, P300, PCAF, GCN5
Total cell lysates were analysed in western blot.
Fig.1
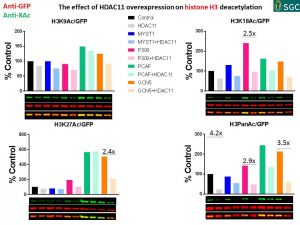
Fig.2
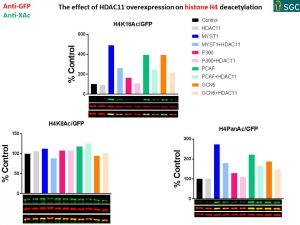
Fig.3
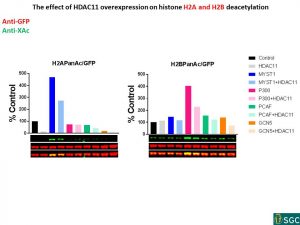
SUMMARY
- The biggest effect of HDAC11 overexpression on histones acetylation:
no HAT PanAc – 4.2x
GCN5 PanAc – 3.5x
p300 PanAc – 2.9x
p300 H3K18Ac – 2.5x
GCN5 H3K27Ac – 2.4x
- HAT acetylation specificity on exogenous histones in Western blot:
MYST1: H4K16 and H2A
P300: H2A>H3K18>H3K27
PCAF: H3K27>H4K16>H3K9,H3K18,H2B
GCN5:H3K27>H4K16>H3K9,H3K18,H2B
You can find the experimental details at https://zenodo.org/record/3460636#.XYto0C5Kipo.
