One of the big aims of the M4K project was to look at the structure of potentially interesting molecules bound to the protein ALK2 and use that to try and design better molecules that might be useful in the treatment of the rare childhood brain cancer, DIPG. Ros did a lot of work on this project and documented a lot of it on her blog. Although currently this project has moved beyond the structure stage, there are still quite a lot of structures that need the final step of tidying up and then depositing into the PDB.
In the last couple of weeks among the other things I’ve needed to do, I’ve also managed to deposit the first of these structure – ALK2 bound to M4K2117.
This compound showed good potency as has been tested by Jong Fu and is one of the more promising compounds (of which there are around 5) with regards to both potency and selectivity for taking forward to the next stage of the process for more in-depth and extensive testing in both cell and animal models.
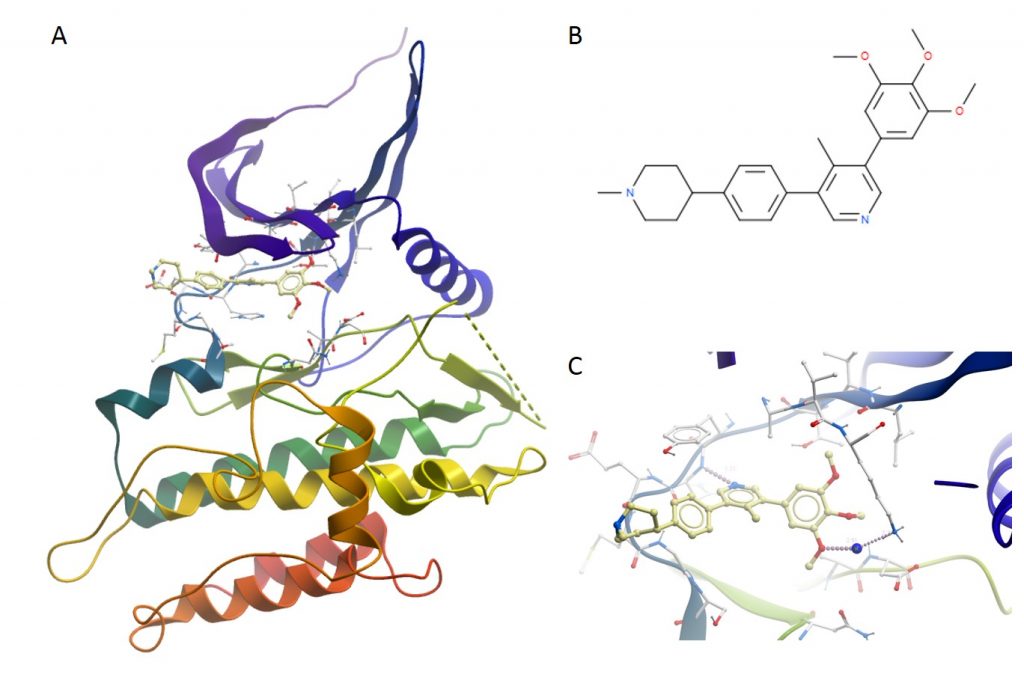
A: The overall structure of ALK2 bound to M4K2117. B: The structure of the ligand M4K2117. C: A close up of the binding pocket of ALK2 bound to M4K2117 showing a standard H bond between the Nitrogen on the central ring and the backbone hinge region as well as a water mediated H bond between a methoxy group on the compound and a side chain at the back of the pocket of ALK2.
The final tweaks to the structures were made using Phenix Refine and Coot while I used the standard wwPDB One Dep system and this structure has been assigned the code: 6SRH
The files for the deposition can be found over on Zenodo and of course you can download the PDB from the PDB website.
