Hello folks, another day another deposition. That makes it sound horribly commonplace but actually Ros put in immense effort into crystallising and solving these proteins and it’s my job just to add the final polish (fix those last stubborn geometry errors), give it a once over to make sure there’s nothing obviously wrong, and whip it into shape to put into the PDB.
This one is another of the M4K structures, this time with M4K2009 which is one of our most promising candidates as shown by Jong Fu and colleagues and is one of the ones being taken forward into more tests. There were two molecules of ALK2 in the asymmetric unit
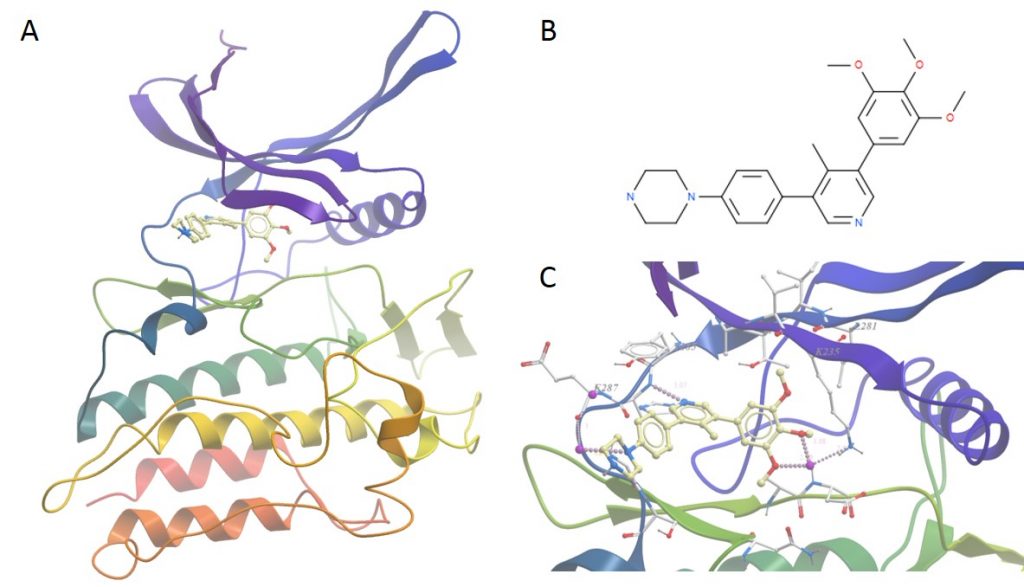
A: Overall structure of ALK2 bound to M4K2009. B: Details of the molecule M4K2009. C: Closeup of the binding pocket showing water mediated H bonds between molecule and side-chains and directly between M4K2009 and the protein hinge region.
The structure is what we expect in a sense, the overall structure of the kinase domain is very similar to usual. Of most interest is the position of water molecules in the binding pocket that help mediate contacts between the protein and the ligand – these help improve the stability and the fit of the compound in the binding site
One other thing of note was the presence of two additional ligand molecules bound to the top of one of the molecules in the asymmetric unit. It is unlikely that these are physiologically relevant and are most likely there as a crystallographic artefact as they are making good contact with chains from the neighbouring unit cell in the crystal.
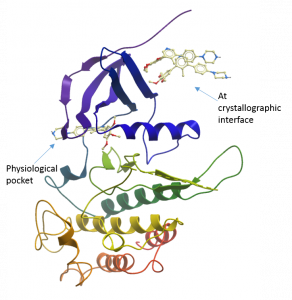
Structure of one copy of ALK2 in the asymmetric unit showing two additional M4K2009 compounds bound non-physiologically at the crystallographic interface.
I used the standard wwPDB One Dep system and this structure has been assigned the code: 6SZM and should be released into the wild (that is, once it has been curated by the people at the pdb) asap!
The files for the deposition can be found over on Zenodo.
