In a previous post, I described the expression and purification of full length (FL) USP5 constructs and also determined that zinc finger ubiquitin-binding (ZnF-UBD) ligands are able to displace ubiquitin in both the ZnF-UBD and the FL protein in a fluorescence displacement assay.
In today’s post, I describe using a surface plasmon resonance (SPR) assay to determine if the ZnF-UBD ligands bind to FL USP5. You can see details of these experiments on Zenodo.
I tested ligands against 4 protein constructs:
- ZnF-UBD wild-type (WT)
- ZnF-UBD R221A
- FL USP5 WT
- FL USP5 R221A
If you’ll recall, R221 is an important residue in the binding pocket of the ZnF-UBD and is involved in ligand co-ordination. A mutation of the arginine (R) to alanine (A) should prevent binding of ubiquitin and the inhibitors to the ZnF-UBD.
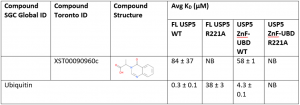
Ligand binding to the WT ZnF-UBD and the FL protein are comparable. For example, in this experiment, compound XST00090960c binds to the ZnF-UBD with a KD of 58 ± 1 µM and to FL USP5 with a KD of 84 ± 37 µM (Table 1). There was no binding of XST00090960c to the ZnF-UBD R221A and FL USP5 R221A confirming that the ligand is indeed binding to the ZnF-UBD in the context of the FL protein. These results corroborate previous displacement assay findings.
Interestingly, ubiquitin binds to FL USP5 R221A (KD=38 ± 3 µM) 100-fold weaker than the FL USP5 WT (KD=0.3 ± 0.1 µM). This is because the FL USP5 contains multiple ubiquitin associated domains that are still able to bind ubiquitin; however the decrease in ubiquitin binding with a R221A mutation shows that the ZnF-UBD is an important domain in co-ordinating ubiquitin binding to the FL protein.
Table 1. SPR data of ZnF-UBD ligands