Today’s post describes testing if ZnF-UBD ligands are able to antagonize USP5 catalytic activity using a ubiquitin-rhodamine110 (UbRho110) assay, which was developed and optimized by Leanna Smith of the Biophysics group at SGC Toronto. You can see details on Zenodo.
The UbRho110 assay is a fluorescence intensity assay that monitors the enzymatic activity of deubiquitinating proteases using a fluorogenic rhodamine-based substrate1. Cleavage of UbRho110 by a deubiquitinase (DUB) enzyme results in monosubstituted rhodamine which exhibits fluorescence when excited at 485 nm1 (Figure 1).

Figure 1. UbRho110 Assay
Introduction of a ZnF-UBD mutant, R221A, described in previous posts, results in approximately 40% DUB activity compared to the WT USP5 (Figure 2). This result is consistent with previous reports which suggest binding of ubiquitin to the ZnF-UBD allosterically activates USP5 catalytic activity2,3. USP5 is a cysteine protease, and cleaves polyubiquitin chains at the site of isopeptide bonds between the C-terminal glycine of one ubiquitin and a terminal nitrogen group of the adjacent ubiquitin. Mutation of the catalytic cysteine (C) to an alanine (A) [C335A] results in no USP5 DUB activity, as expected.
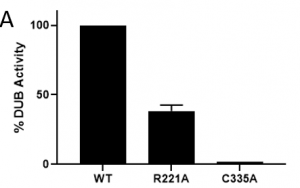
Figure 2. DUB activity of USP5 constructs (n=2)
To determine if ZnF-UBD ligands antagonize USP5 activity in the UbRho110 assay, I tested one of our most promising ligands, XST00090960c using optimized assay conditions. XST00090960c inhibits approximately 75% of USP5 activity at the maximum concentration tested (Figure 3). Therefore, ZnF-UBD ligands are able to antagonize USP5 catalytic cleavage of the UbRho110 substrate. It should be noted, UbRho110 is not a native substrate, and further validation with native polyubiquitin species in an orthogonal assay is required. In the future I plan to use a western blot assay to determine whether ZnF-UBD ligands antagonize native polyubiquitin species.
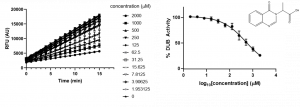
Figure 3. Representative activity data of compound XST00090960c
References
1Hassiepen, U.; Eidhoff, U.; Meder, G.; Bulber, J. F.; Hein, A.; Bodendorf, U.; Lorthiois, E.; Martoglio, B. A Sensitive Fluorescence Intensity Assay for Deubiquitinating Proteases Using Ubiquitin-Rhodamine110-Glycine as Substrate. Anal. Biochem. 2007, 371, 201-207.
2Reyes-Turcu, F. E.; Horton, J. R.; Mullally, J. E.; Heroux, A.; Cheng, X.; Wilkinson, K. D. The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin. Cell 2006, 124, 1197-1208.
3Avvakumov, G. V.; Walker, J. R.; Xue, S.; Allali-Hassani, A.; Asinas, A.; Nair, U. B.; Fang, X.; Zuo, X.; Wang, Y. X.; Wilkinson, K. D.; Dhe-Paganon, S. Two ZnF-UBP Domains in Isopeptidase T (USP5). Biochemistry 2012, 51, 1188-1198.
