Background
RFWD3 (E3 ligase RING finger and WD repeat domain-containing protein 3) and RPA2 (Replication protein A2) were shown to functionally interact and participate in replication checkpoint control. At stalled replication forks, the RPA complex binds single-stranded DNA to prevent nucleolytic digestions and also to initiate replication checkpoint signaling by recruitment checkpoint proteins. RFWD3 is recruited to stalled replication forks and interacts directly with RPA2 in response to replication stress. The deletion of the RPA2 binding region on RFWD3 impairs its localization to stalled replication forks (PIMID: 21504906).
Summary
Assay measuring the interaction of RPA2 to RFWD3 was developed using NanoBRET technology – a proximity-based assay that can detect protein interactions by measuring energy transfer from a bioluminescent protein donor to a fluorescent protein acceptor. We determined the optimal donor to acceptor ratio and levels of expression of 2 donor constructs: C- or N-terminally NanoLuc®(NL)- tagged RFWD3 and acceptor construct: N-terminally HaloTag®Fusion (HT)-tagged RPA2. We compared the results to background NanoBRET signal from HT-RPA2 and NL protein alone. The best results were obtained with N-terminally NL-tagged RFWD3 (0.001 µg/96-well) and N-terminally HT- tagged RPA2 (0.03 µg/96-well) (Fig.1)
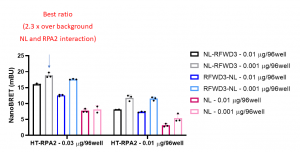
Fig 1. RPA2 – RFWD3 nanoBRET assay. HEK293T cells were plated in 96-well plates and transfected with indicated amounts of C- or N- terminally NL-tagged RFWD3 and N-terminally HT-tagged RPA2. Next day cells were treated +/- fluorescent ligand and 4 h later the luciferase substrate was added, and the signal was read. The results are mean +/- SD of 3 replicates.
Go to Zenodo for experimental details.
