Background:
Scientists centred around University of Oxford came together in the current COVID-19 pandemic to contribute to relevant studies collaboratively. Researchers with no prior experience in virology studies, myself included, had volunteered our time to contribute whichever way we can based on our field of expertise. The Biotech team of SGC Oxford, headed by Nicola Burgess-Brown had taken up the role of producing proteins used in serological assays, basic research to understand COVID-19 pathophysiology and hunt for treatment compounds. The initial production of these proteins was based on transient transfection of expression construct DNA into mammalian cells. This approach was laborious and consumed large amounts of plasmid DNA. Therefore, I volunteered to generate stable expression cell lines that will negate the constant need for transfection.
Experimental design:
I aimed to stably integrate the expression constructs into the genome of Expi293F human cells commonly used in large scale protein production. The expression constructs carried neomycin marker for selection in mammalian cells. Toxicity to the cells is not a concern in this case because the expressed proteins are secreted into the medium and do not build up in the cells. The expression constructs are randomly integrated into the genome mainly via non-homologous end-joining (NHEJ). An advantage of this approach is that molecular sub-cloning is not necessary.
In order to maximise the rate of correct integration of the expression constructs, restriction digestion was performed to cut once between the expression cassette and antibiotic resistance cassette (refer to cartoon in Figure 1). Without this step, the construct DNA will be broken at a random position during the integration step, potentially disrupting the expression cassette. I have also tested the effect of actively creating double strand breaks (DSBs) in the genome of the cells on the integration efficiency. More DSBs in the genome might increase the incidence of NHEJ and integration of our constructs. This was achieved by co-transfection of AAVS1 CRISPR/eSpCas9(1.1) construct (refer to cartoon in Figure 1). The eSpCas9(1.1)_No_FLAG_AAVS1_T2 construct was a gift from Yannick Doyon (Addgene plasmid # 79888 ; http://n2t.net/addgene:79888 ; RRID:Addgene_79888)
Results:
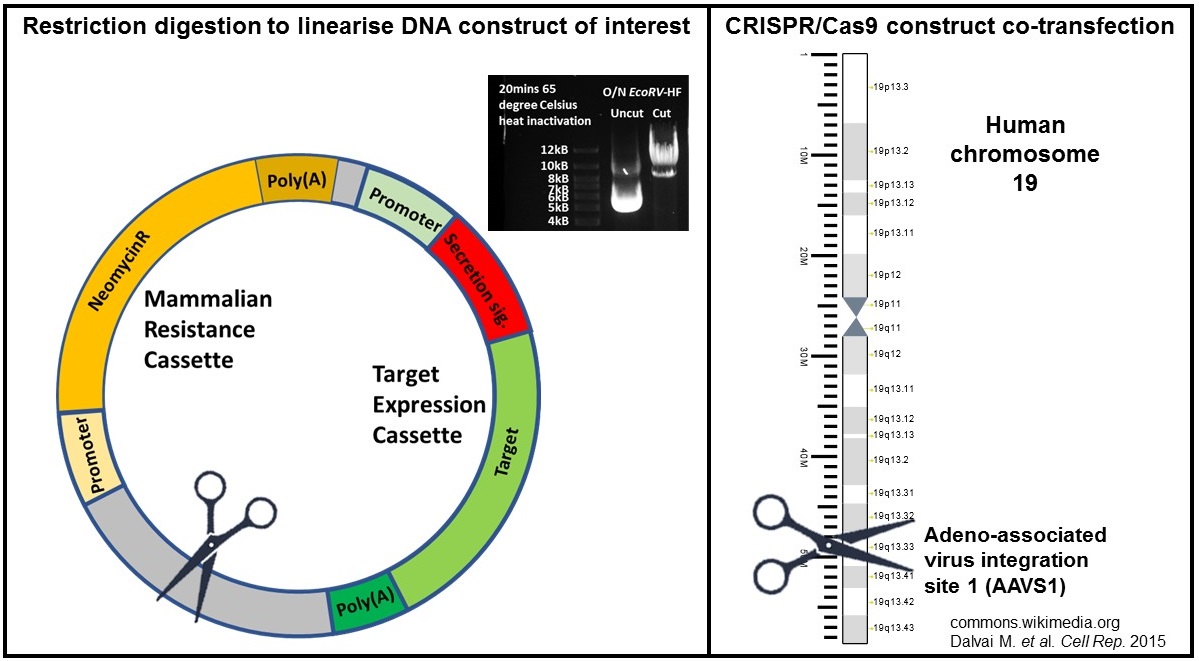
Cartoons of approaches to enhance desired stable integration of protein expression constructs.
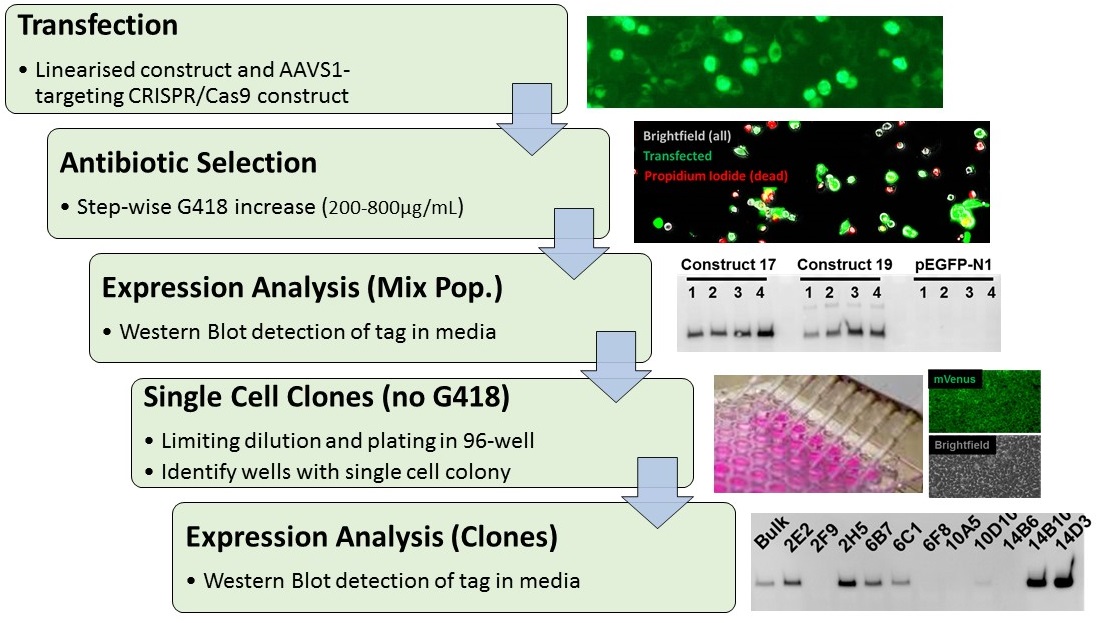
Schematic of the workflow to generate stable cell lines
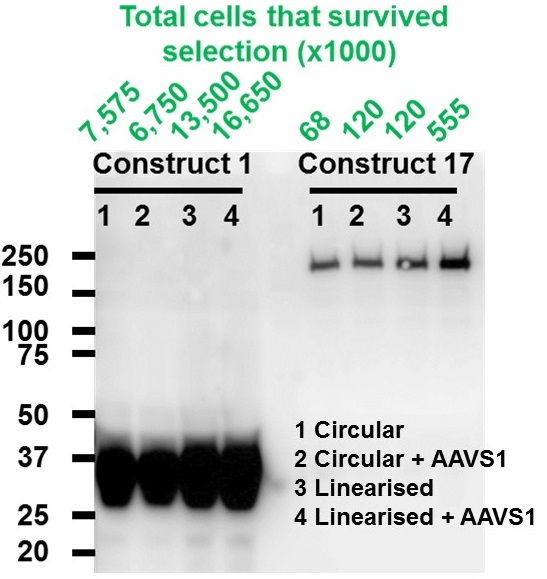
Comparison of the efficiency of correct stable integration under different conditions. Numbers of surviving cells were counted after G418 antibiotic selection and the expression of desired proteins was detected using Western Blot.
Findings:
- Linearisation of expression construct increased stable integration.
- Induction of AAVS1 double strand break enhanced stable integration further.
- The efficiency of stable integration is highly dependent on construct size. E.g. 12.3 kb construct is about 30 times less efficiently integrated than 8.2 kb construct.
- More than 5-fold increase in purified proteins compared to same volume of transient transfected cells.
In a nut shell:
Stable cells that continuously produce and secrete high level of COVID-19-related proteins have been successfully generated. These cells are suitable for large scale culture and protein purification. These stable cells will help to streamline protein production for serological assays and other COVID-19-related studies.
For detailed protocol and information, please refer to my Zenodo post.
