In my previous post 19, I showed how we assessed the druggability of the remdesivir-binding site of SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) and determined the residues lining that site. In this post, we analyze the genetic diversity of RdRp catalytic site across Alpha- and Betacoronavirus entries from UniProt and among SARS-CoV-2 samples.
In the context of the emergence of future MERS-like or SARS-like coronaviruses from the bat strains circulating in bat reservoir species, it is imperative to do a broad survey of viral proteins to identify the best strategies for the development of broad-spectrum viral inhibitors. (Sheahan et al., 2020) For this exact reason, we are looking at twenty-seven reviewed sequences from Uniprot, where six belong to entries of the Alphacoronavirus genus and twenty-one belong to the Betacoronavirus genus. We then assess the variability of amino acid lining the binding pockets found at the RdRP active site (PDB: 7bv2).
I made a diversity dendrogram focusing on the 48 amino acid sidechains lining the pockets juxtaposed to Remdesivir. Shown below in figure 1, the consensus profile at each of the 48 sidechains is shown. Strikingly, 83% of residues, 40 out of 48, are conserved across these 27 entries. This is one of the most conserved SARS-CoV-2 binding sites that we have seen so far (I will post a systematic comparison once I have reviewed all sites on all SARS-CoV-2 proteins in the PDB). Here are the 8 non-conserved residues: F442, A443, Y453, Y455, M543, N552, A558, S681. Across the 21 entries of the Betacoronavirus genus (including SARS-CoV-2), 46 out of 48 residues are conserved at the RdRp active site. Here are the non-conserved residues: F442 (almost perfectly conserved), and A443.
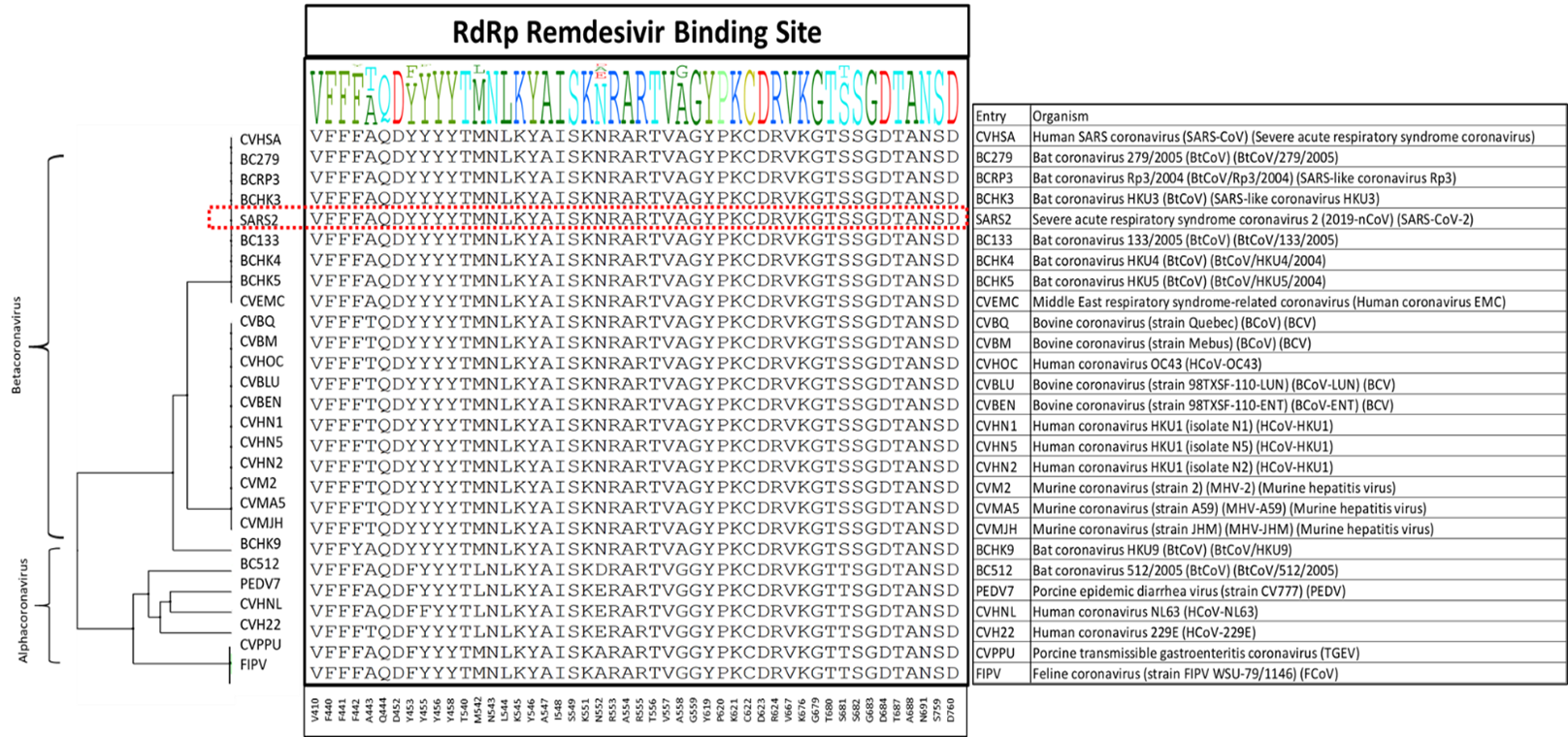
Figure 1. The diversity dendrogram of the active site of RdRp for the entries of Alpha- and Betacoronavirus genera.
We mapped the variations shown among the entries of Alpha- and Betacoronavirus genera onto the SARS-CoV-2 RdRp crystal structure (PDB: 7bv2) such that sidechains that are variable among the entries are highlighted in orange and the conserved sidechains are shown in green.
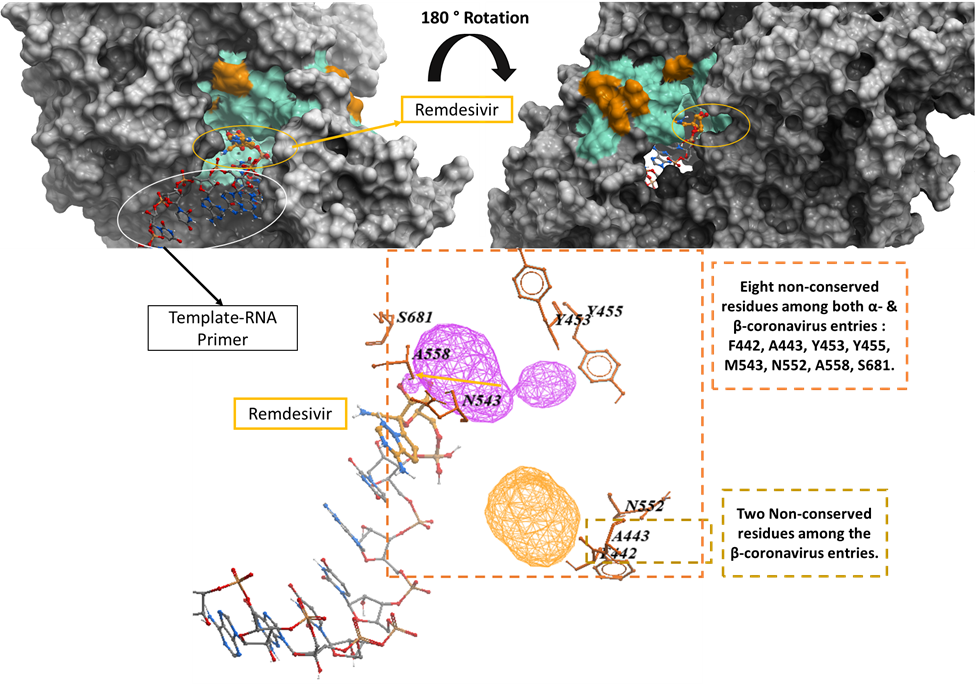
Figure 2. Mapping the genetic variation of UniProt entries belonging to Alpha- and Betacoronavirus genera onto SARS-CoV-2 RdRp Crystal structure (PDB: 7bv2). The eight non-conserved residues among the entries of both Alpha- and Betacoronavirus include F442, A443, Y453, Y455, M543, N552, A558, S681 while the two non-conserved residues among the Betacoronavirus entries include F442, A443.
Our next step was to assess the genetic variability of the catalytic site of RdRp among SARS-CoV-2 samples. Nicola De Maio, our collaborator from Nick Goldman’s lab at European Bioinformatics Institute, looked at more than 15000 SARS-CoV-2 samples and identified all mutations at the RdRp catalytic site pockets. He identified 6 non-synonymous variants as shown in table 1. For example, at residue number 443, the wild-type sidechain in SARS-CoV-2 RdRp is alanine. In the sample batch Nicola has looked at, he saw that 15870 samples had alanine at that position while there were 2 samples with valine at this exact position.
| Index | Non-synonymous Variants | variants | codon one | codon two | codon three |
| 1 | A443 | A(15870), V(2) | g(15872) | c(15870), t(2) | t(15872) |
| 2 | Q444 | Q(15870), H(2) | c(15872) | a(15872) | t(2), g(15870) |
| 3 | M542 | M(15873), L(1) | a(15874), c(1) | t(15875) | g(15874) |
| 4 | A547 | A(15874), V(1) | g(15875) | c(15874), t(1) | c(15875) |
| 5 | A554 | A(15874), V(1) | g(15875) | c(15874), t(1) | c(1), t(15874) |
| 6 | V667 | V(15870), I(5) | a(5), g(15870) | t(15875) | c(15875) |
Table 1. The non-synonymous variants at RdRp active site across more than 15000 SARS-CoV-2 samples.
We could not predict the effect of these mutations on remdesivir binding due to the lack of RdRp structure where remdesivir has not been incorporated into the RNA primer sequence while it is sitting in a pocket in RdRp. Therefore, we can only comment on how the nature of the variant sidechains differ from the wild-type sidechains according to their physiochemical properties: the size and polarity of mutated residues are largely conserved in all cases (A->V, Q->H, V->I), suggesting high chances of a light effect on ligand binding.
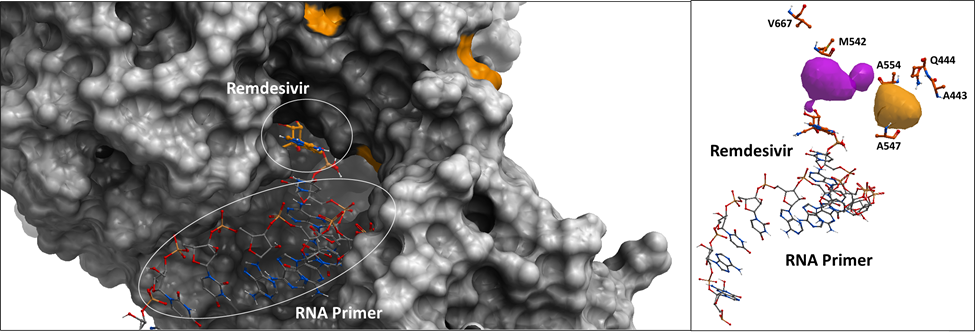
Figure 2. Left Panel: The surface representation of SARS-CoV-2 RdRp crystal structure with the non-conserved amino acid positions highlighted in orange. Right Panel: The sticks representation of the non-conserved residues among SARS-CoV-2 samples.
In conclusion, from a structural standpoint, the catalytic site of RdRp is a good target for broad-spectrum inhibitors against coronaviruses considering that only 17% of the residues lining catalytic site binding pockets were not conserved across 27 Alpha- and Betacoronavirus entries. It is worth mentioning that clinically, the inhibition of RdRp which is achieved by remdesivir has not been fully confirmed to be an effective therapy for COVID-19.(Goldman et al, 2020)
According to the SARS-CoV-2 variants reported in table 1, the properties of most of the non-synonymous variants at the catalytic site were conserved, suggesting high chances of a light effect on ligand binding.
It would be crucial to investigate the structure of RdRp combined with remdesivir in the state right before it gets incorporated into the RNA primer. This would reveal the main interactions that remdesivir makes with RdRp sidechains and allow to better predict the effect of mutations both across coronaviruses broadly and more specifically across samples from COVID-19 patients on remdesivir binding.
To view the full report on SARS-CoV-2 RdRp, please visit this Zenodo report.
As always, I would be happy to hear your comments. Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for my next post!
References:
Sheahan, T. P., Sims, A. C., Zhou, S., Graham, R. L., Hill, C. S., Leist, S. R., Schäfer, A., Dinnon, K. H., Montgomery, S. A., Agostini, M. L., Pruijssers, A. J., Chapell, J. D., Brown, A. J., Bluemling, G. R., Natchus, M. G., Saindane, M., Kolykhalov, A. A., Painter, G., Harcourt, J., … Baric, R. S. (2020). An orally bioavailable broad-spectrum antiviral inhibits SARS-Cov-2 and multiple endemic, epidemic and bat coronavirus. https://doi.org/10.1101/2020.03.19.997890
Goldman, J. D., Lye, D. C.B., Hui, D. S., Marks, K. M., Bruno, R., Montejano, R., Spinner, C. D., Galli, M., Ahn, M., Nahass, R. G., Chen, Y., SenGupta, D., Hyland, R. H., Osinusi, A. O., Cao, H., Blair, C., Wei, X., Gaggar, A., Brainard, D. M., & Subramanian, A. (2020). Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. The New England Journal of Medicine. https://doi.org/10.1056/NEJMoa2015301
