Previously we reported our analysis on the structural diversity of binding pockets found on the SARS-CoV-2 main protease, methyltransferase, and papain-like protease, macrodomain. We then shifted our focus on the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp).
SARS-CoV-2 is a positive-strand RNA virus, depending on its multi-subunit machinery to replicate its RNA. The catalytic subunit of RdRp is the core component of this machinery, which is also known as nsp12. Nsp12 relies on accessory subunits to have full activity. The accessory factors, nsp7 and nsp8, increase RdRp template binding and processivity. (Yin et al., 2020)
RdRp is a well-established target for a class of antiviral drugs known as nucleotide analogs. One of the known and only FDA and Health Canada approved medications for COVID-19, remdesivir, targets RdRp.
Remdesivir is converted to its active form, remdesivir triphosphate (RTP), that gets incorporated into RNA and causes chain termination.
It is important to note that remdesivir has been tested in phase III clinical trial. The results were shared in The New England Journal of Medicine on May 27, 2020, by Goldman and his colleagues. (Goldman et al, 2020) The use of remdesivir for 5 to 10 days in patients with severe COVID-19 showed that there was no significant difference between the 5-day treatment and 10-day treatment groups. The overall effect on the survival rate was rather low. It is important to note this study did not have a placebo control arm and that means the magnitude of the benefit cannot be determined.
Yin et al solved the crystal structure of RdRp in complex with a remdesivir-incorporated RNA strand (PDB code 7bv2). The authors published two structures of RdRp; one in the apo form containing one nsp12, nsp7, and two nsp8 proteins (PDB: 7bv1) shown in figure 1A and another bound to the remdesivir-incorporated RNA, containing nsp12, one nsp7, and one nsp8 (PDB: 7bv2) shown in figure 1B.
Upon incorporation of the remdesivir into the primer strand, there is immediate termination of chain elongation. This inhibitory mechanism is called nonobligate RNA chain termination by remdesivir where the prodrug needs conversion to the active triphosphate form, RTP. (Yin et al., 2020)The interactions between remdesivir monophosphate (RMP) and RdRp are shown in figure 2.
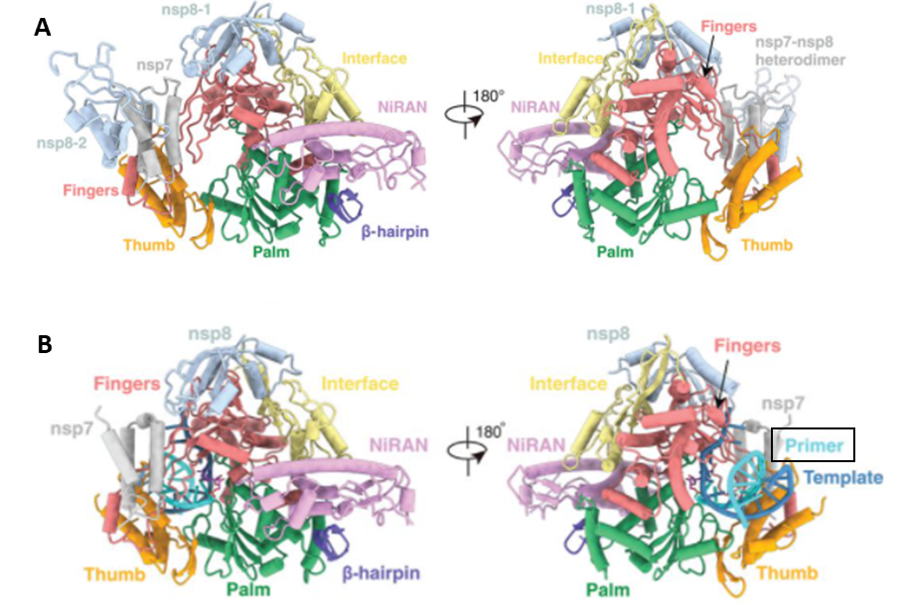
Figure 1. (A) Views of Cryo-electron microscopy structure of nsp12-nsp7-nsp8 RdRp complex (B) and nsp12-nsp7-nsp8 in complex with the primer RNA template and remdesivir (Yin et al., 2020)
Once RdRp is in the template-RTP form, it goes into a closed conformation where the 11 base-pairs template primer is held by the finger-palm-thumb subdomains. There are also extensive RNA-protein interactions that involve the RNA phosphate-ribose backbone. In the absence of specific interactions between any specific base-pairs and RdRp, the binding of RNA by RdRp becomes independent of the RNA sequence which explains why no specific sequence is needed for RdRp’s enzymatic activity. (Yin et al., 2020)
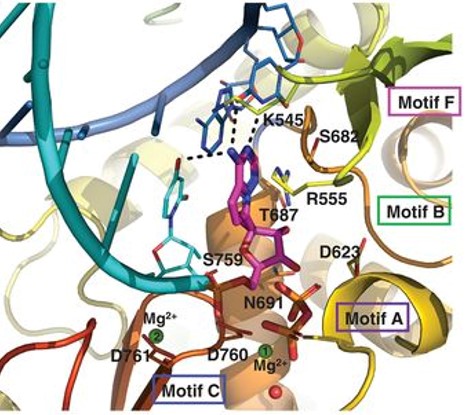
Figure 2. The chemical interactions between the RdRp catalytic site residues and RMP (purple). (Yin et al., 2020)
We aim to analyze the genetic diversity of RdRp catalytic site across coronaviruses and SARS-CoV-2 samples. To achieve this goal, we first assessed the druggability of the remdesivir-binding site of RdRp and determined the residues lining that site which I explain in this post.
Using SiteMap (Schrodinger, NY) we assessed the druggability of the binding sites using the druggability score (Dscore). We found that the active site of RdRp in the vicinity of bound remdesivir has a Dscore of 0.859 indicative of a difficult druggable site. We show a schematic of this site in figure 3.
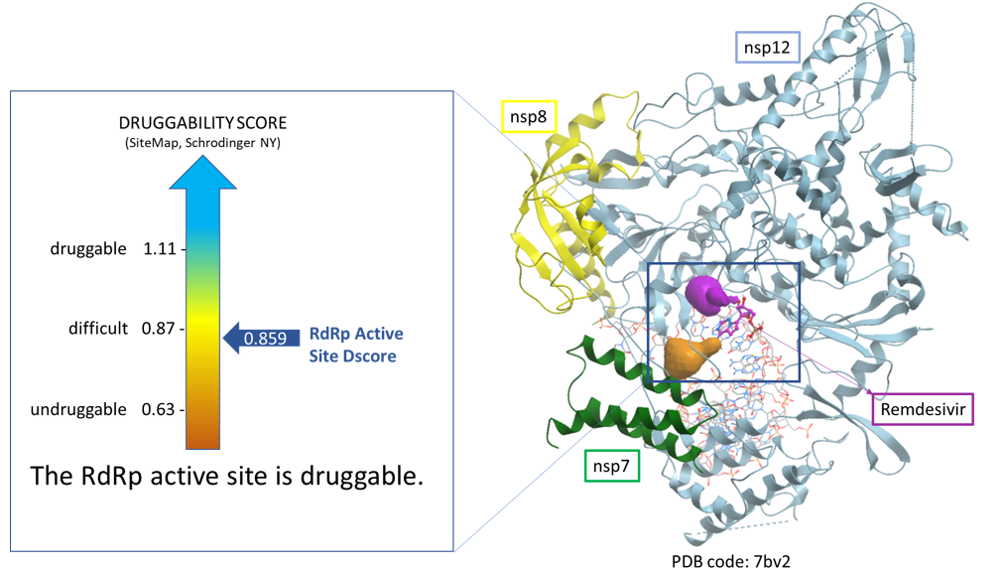
Figure 3. Druggability analysis of SARS-CoV-2 RdRp active site. Cavities at the catalytic site (orange and magenta blobs) are found next to the position occupied by remdesivir monophosphate incorporated into the RNA template (purple stick).
Using ICM PocketFinder, we find that RMP is not occupying a clearly defined binding pocket but is juxtaposed to two cavities. This probably reflects the fact that we are looking at a structure where remdesivir was already incorporated into RNA. The site is structurally conserved in the apo form (PDB: 7bv1). We hypothesize that compounds occupying these cavities would block the catalytic activity of RdRp. We identified 48 amino acid sidechains lining the pockets juxtaposed to Remdesivir shown in figures 3 and 4 (within 2.8 Å of the pocket): V410, F440, F441, F442, A443, Q444, D452, Y453, Y455, Y456, Y458, T540, M542, N543, L544, K545, Y546, A547, I548, S549, K551, N552, R553, A554, R555, T556, V557, A558, G559, Y619, P620, K621, C622, D623, R624, V667, K676, G679, T680, S681, S682, G683, D684, T687, A688, N691, S759, D760.
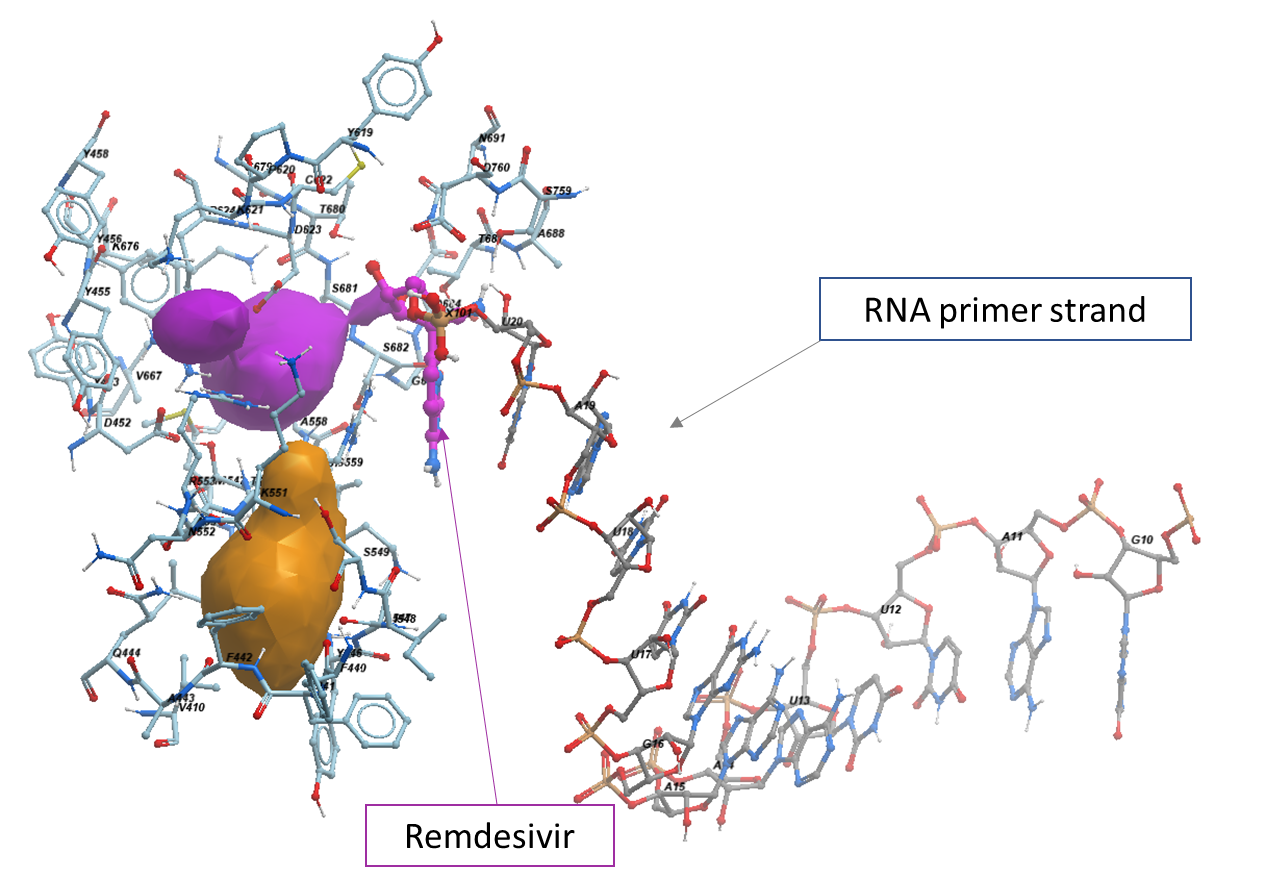
Figure 4. The neighboring residues lining the active site of RdRp.
In my next post, I will show the sequence diversity across UniProt entries from the Alpha- and Betacoronavirus genera and map that to the RdRp remdesivir-binding site using its crystal structure.
As always, I would be happy to hear your comments. Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for my next post!
References:
Yin, W., Mao, C., Luan, X., Shen, D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y., Tian, G., Jiang, H., Tao, S., Shen, J., Jiang, Y., Jiang, H., Xu, Y., & Xu, H. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science, 368(6498), 1499-1504. https://doi.org/10.1126/science.abc1560
Goldman, J. D., Lye, D. C.B., Hui, D. S., Marks, K. M., Bruno, R., Montejano, R., Spinner, C. D., Galli, M., Ahn, M., Nahass, R. G., Chen, Y., SenGupta, D., Hyland, R. H., Osinusi, A. O., Cao, H., Blair, C., Wei, X., Gaggar, A., Brainard, D. M., & Subramanian, A. (2020). Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. The New England Journal of Medicine. https://doi.org/10.1056/NEJMoa2015301
