Production and purification of Ectodomain of SARS-CoV-2 Spike (S) protein
Two different expression plasmids for prefusion S ectodomain residues 1−1208 of 2019-nCoV S (GenBank: MN908947) with proline substitutions (2P) at residues 986 and 987(1) and with six proline substitutions (HexaPro (at residues 817, 892, 899, 942, 986 and 987 (2), a “GSAS” substitution at the furin cleavage site (residues 682–685), a C-terminal T4 fibritin trimerization motif, an HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag in the mammalian expression vector pαH were generously gifted by Dr.McLellan’s lab (1,2).
I. Production and purification of the 2P version of S-Ectodomain.
During the first purifications from a small-scale production we have faced some challenges with the 2P version of this protein. (Figure 1.)
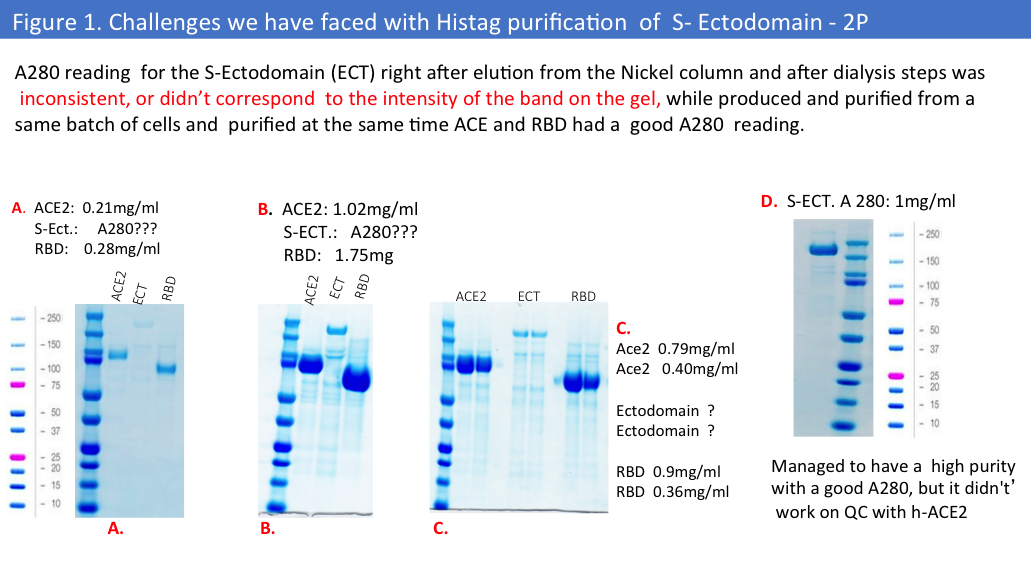
Purification from a big scale-up production using another buffer (1x PBS) led to the same problems, and we evaluated protein concentrations by Pierce BCA Protein Assay (Please see Figure 2&3) and comparative gel analysis.
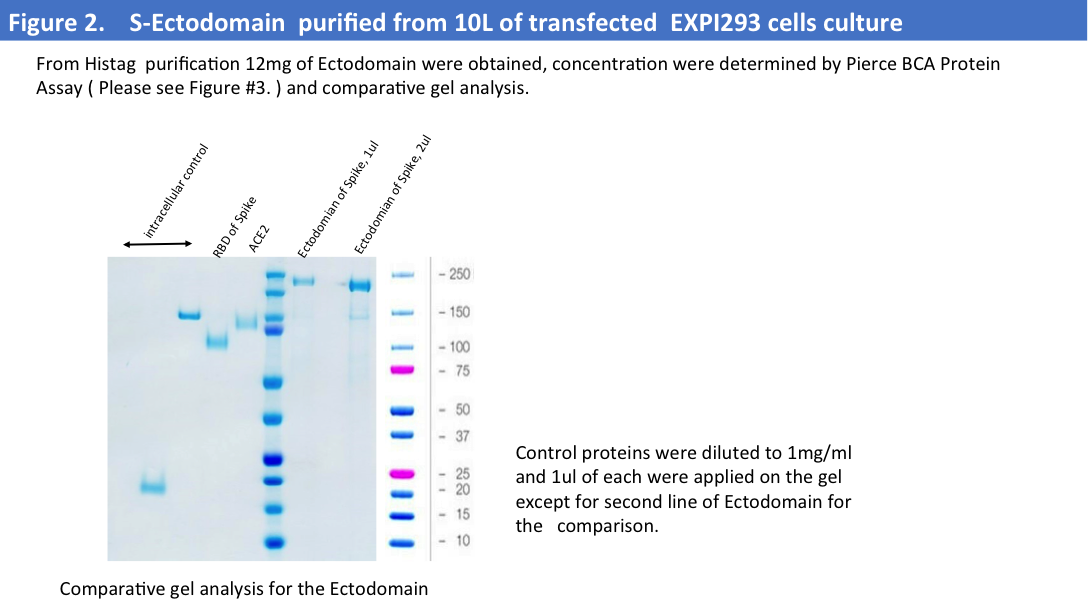
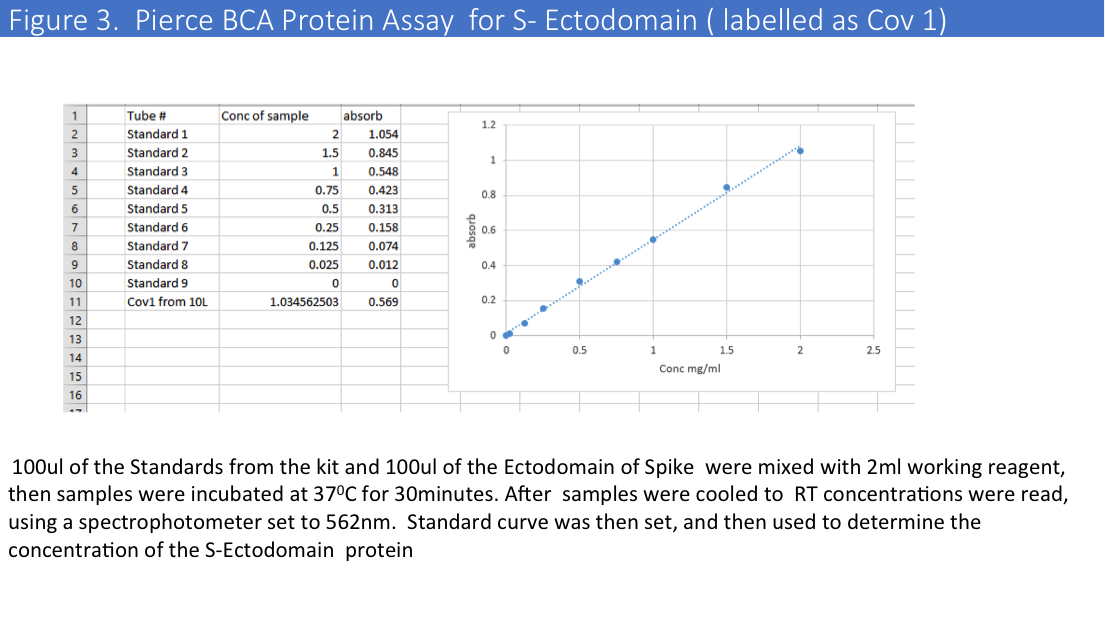
We have obtained total 12mg of Histag purified and dialyzed protein and run 9mg of protein on the size-exclusion column. SEC profiles showed that a big portion of the protein were in aggregates (Figure 4).
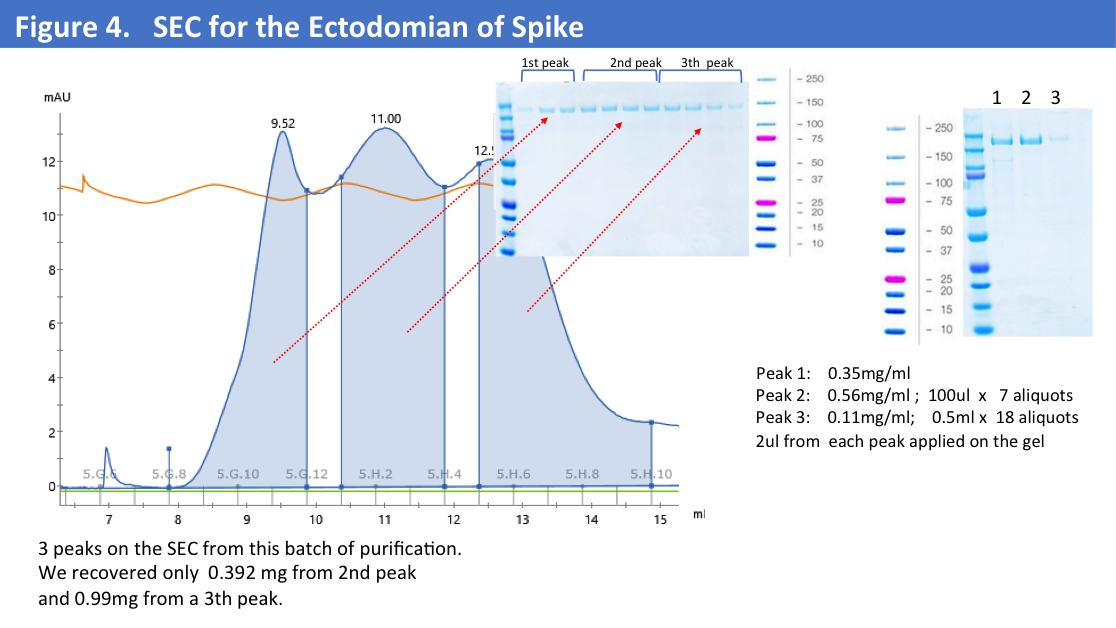
On the size-exclusion profile we observed 2 peaks of the aggregates and collected only a 3rd peak corresponding to the trimer of S- Ectodomain running close to 670kD.
Based on the results of the purifications by affinity, size-exclusion protocols and exchanging notes with a Biotechnology group of SGC Oxford, we’ve come to conclusions that S- Ectodomain is prone to the aggregation almost at the every stage of purification, one nucleation of the aggregates “ curls up” a most part of the protein.
We have optimised a few steps in the production and purification of this target to prevent or minimize aggregation:
• Cultured cells were harvested in a different hours of the post-transfection times
• Harvested culture supernatant clarified by two steps of centrifugation at RT was passed through a
0.22 μm filter before IMAC batch absorption
• After IMAC purification, combined peak fractions were filtered through a 0.22 μm filter, before
concentrating and loading to the SEC
• Concentrated combined fractions of the eluted protein at 16 degree celsius and run on SEC
Size-exclusion profiles of S-Ectodomain from these 4 purifications shown no major aggregates and a main fraction of the protein was corresponding to the expected MV as 670kD (Figure 5.), but a final yield of the purified by two-steps protein were very low as 0.2-0.3mg/L.
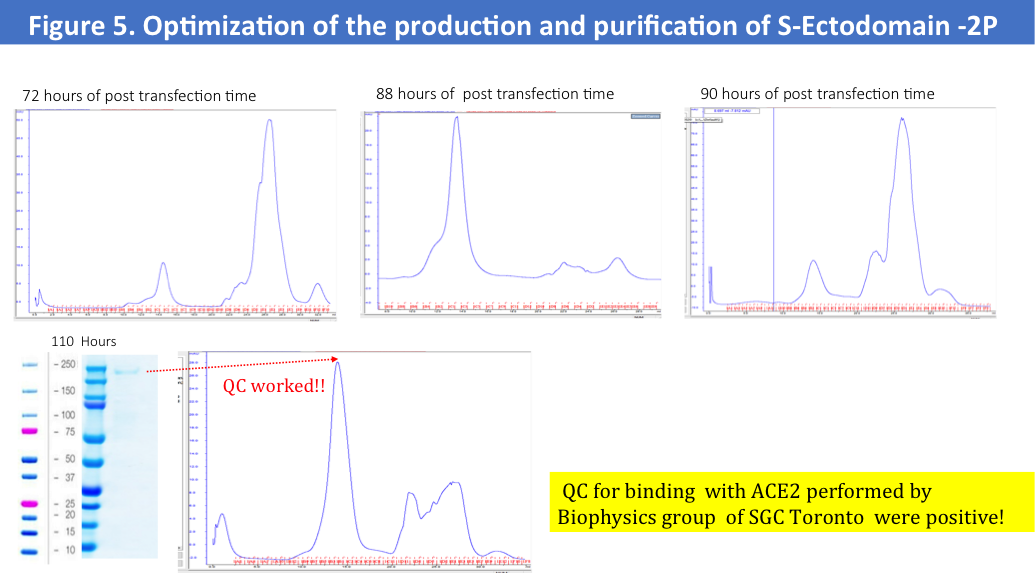
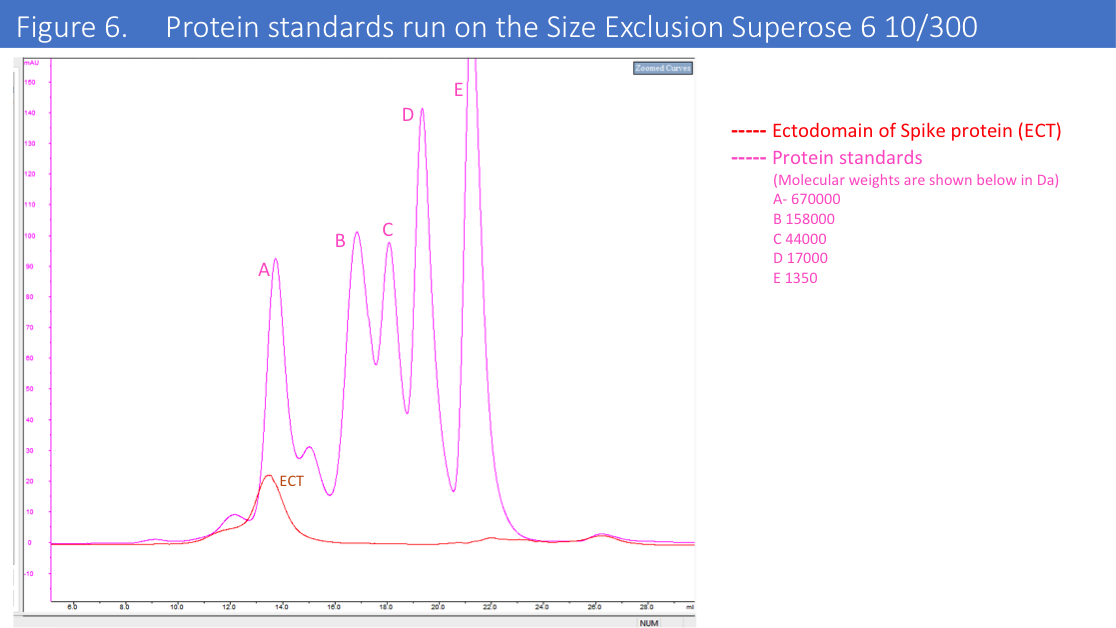
We have run protein standards on the SEC column used for this protein to confirm accuracy of the MV of the eluted protein (Figure 6.)
II. Production and purification of the HexaPro version of S-Ectodomain
We have used the same optimized protocol for the HexaPro version of the S-Ectodomain (Figure 7).
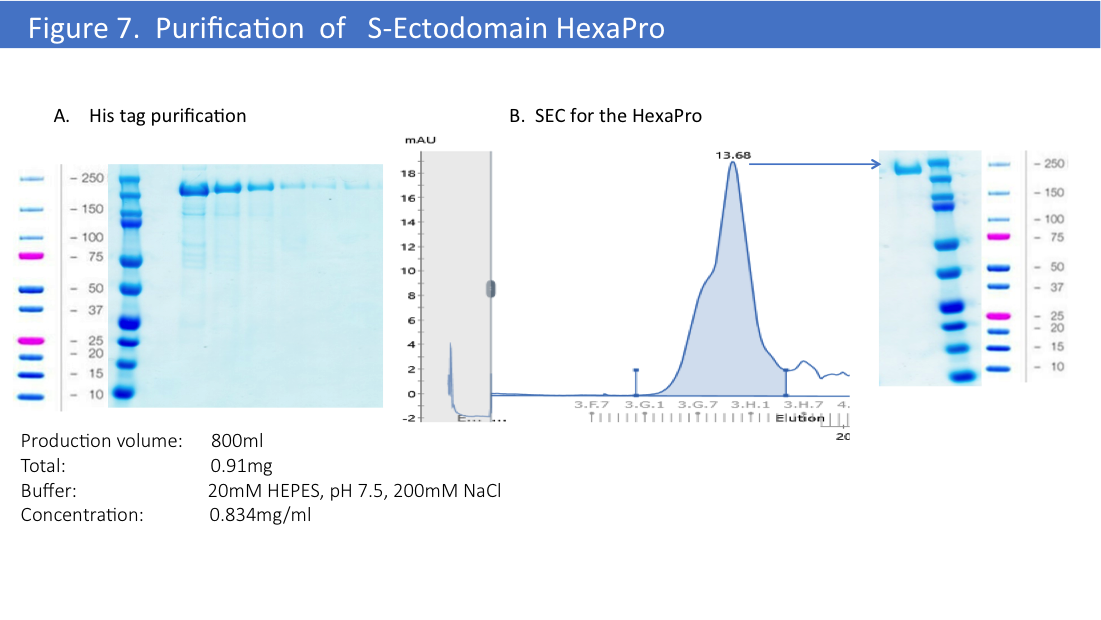
Final yield for this constructs was higher than for 2P version of S-Ectodomain, we’ve obtained 1mg/L from the first 800ml of production by transient transfection of EXPI293 cells. Further optimization might improve a final yield to the higher expression for this construct, but so far we’ve done only one production and purification trial with HexaPro.
References:
1.Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
Daniel Wrapp, Nianshuang Wang, Kizzmekia S. Corbett, Jory A. Goldsmith, Ching-Lin Hsieh, Olubukola Abiona, Barney S. Graham, Jason S. McLellan
Science 367, 1260–1263 (2020) 13 March 2020
2. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes
Ching-Lin Hsieh, Jory A. Goldsmith, Jeffrey M. Schaub, Andrea M. DiVenere, Hung-Che Kuo,
Kamyab Javanmardi, Kevin C. Le, Daniel Wrapp, Alison G. Lee, Yutong Liu, Chia-Wei Chou,
Patrick O. Byrne, Christy K. Hjorth, Nicole V. Johnson, John Ludes-Meyers, Annalee W. Nguyen,
Juyeon Park1, Nianshuang Wang1, Dzifa Amengor1, Jason J. Lavinder1,2, Gregory C. Ippolito,
Jennifer A. Maynard, Ilya J. Finkelstein, Jason S. McLellan
Science 369, 1501–1505 (2020) 18 September 2020
Please see Zenodo page for the production and purification protocols
