In this blog post, I will discuss the progress made on improving the binding affinity of new covalent quinazolines targeting CYS122, as well as some of the directions we are taking to further improve binding affinity to TBXT and reduce binding affinity towards EGFR. The synthetic work has been primarily driven by Zach and our CRO – Piramal, and mass spec labeling and crystallographic data have been performed by Joseph Newman and Nergis Imprachim at Oxford. This post is part 2 of the 2-part series for covalent compounds we have been pursuing.
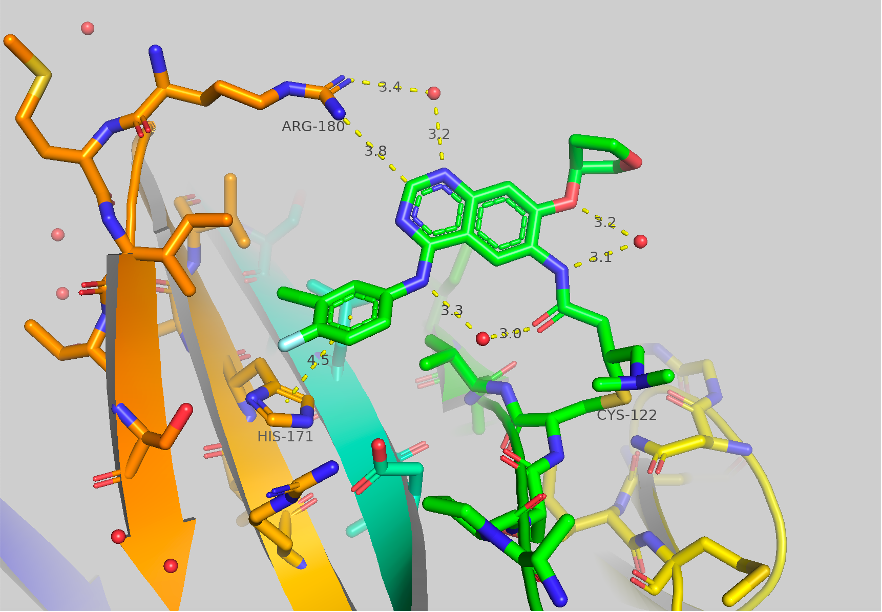
Figure 1 Crystal structure of afatinib bound to TBXT, 6ZU8. Contacts to HIS171 and ARG180 are shown as well as some potentially important bridged water molecules.
The crystal structure of afatinib bound to TBXT is shown in Figure 1. Contacts to HIS171 and ARG180 are shown, although they are quite long. A few potentially significant water molecules are present between the amide warhead and the ortho-ether and the NH aniline. A water molecule is also shown between ARG180 and the 1-N of the quinazoline core. This structure as mentioned in Part A is the starting point for our work.
In our mass spec binding assay, we found that afatinib labels 10% and 72% at 5 mM and 50 mM, respectively, over 1 h. Like our work discussed in Part A: Benzamides we initially targeted the 4-aniline group to pursue the initial SAR for improving binding to brachyury. As seen in Figure 1 there is only a long contact (4.5 Å) made to HIS-171, and we hypothesized that this contact, as well as potential contacts to other neighboring amino acids, could be optimized.
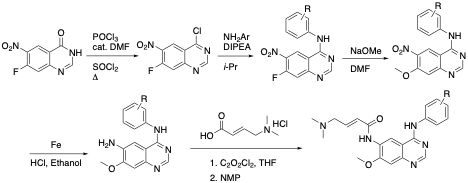
Figure 2 Synthetic strategy for the synthesis of afatinib analogs.
Our synthetic work follows literature-reported procedures for the preparation of covalent quinazolines. Our CRO, Piramal, performed this work as the synthesis required for the preparation of a large SAR library is quite intensive, requiring 4 steps for each aniline (Figure 2). We have prepared and tested 40 different quinazoline derivatives which we have previously shared in our Zenodo data release.
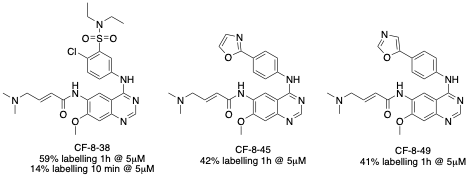
Figure 3 Top hits from first-round afatinib SAR.
During one of our early screenings of afatinib analogs, we identified three compounds that showed significant improvement in labeling compared to afatinib (Figure 3). Surprisingly, the aniline of CF-8-38 is the same aniline that gave major improvements discussed in Part A: Benzamides. The other two compounds (CF-8-45, and CF-8-49) showing major improvement contained two different oxazole isomers in the p-position. We also tested CF-8-38 on a shorter time frame (10 minutes) and found 14% labeling at this time. Lately, our team has run this assay at both time points.
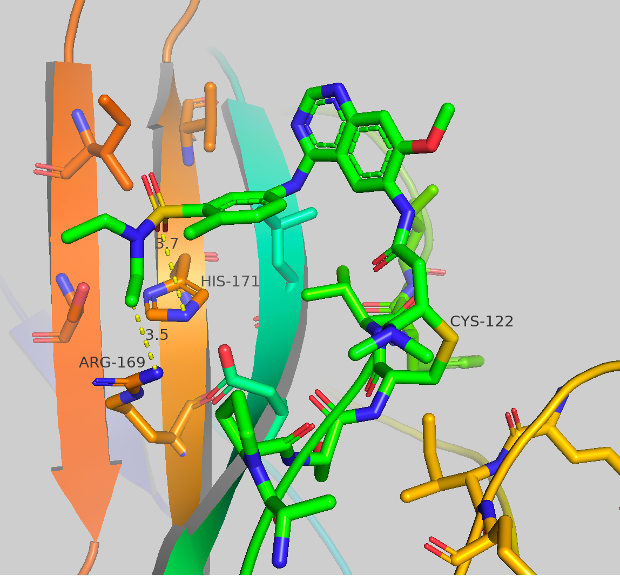
Figure 4a Crystal structures of CF-8-38.
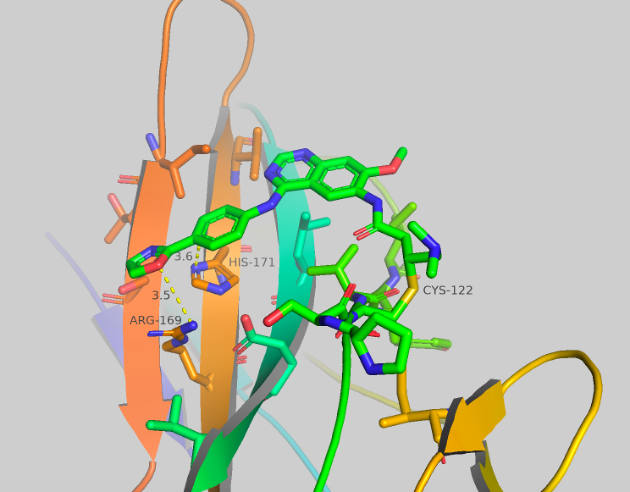
Figure 4b Crystal structures of CF-8-45.
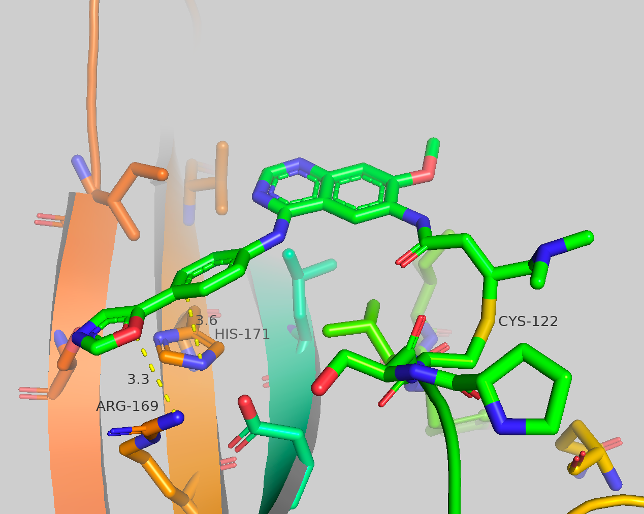
Figure 4c Crystal structures of CF-8-49.
Figure 4a-c Crystal structures of CF-8-38; CF-8-45; CF-8-49. With major contact distances shown to the corresponding amino acids of brachyury.
We were able to generate crystal structures of each of these compounds, shown in Figure 4. The three new compounds show similar crystal structure form and general orientation as afatinib but with some differences. CF-8-38 shows the most similarities to afatinib, with the quinazoline core twisted, nearly perpendicular to the surface of brachyury. The sulfonamide group makes 2 contacts to brachyury, an aromatic-pi interaction to HIS171 from one of the oxygens (3.7 Å) of the sulfonamide, and close contact to ARG169 from the ethyl group of the amine (3.5-3.7 Å). No waters are present which may have an impact on ARG180, as this AA has shown to be extremely flexible. CF-8-45 and CF-8-49 show more significant differences to the afatinib structure. The quinazoline core is now flat and closer to parallel to the surface of brachyury. The contacts made to HIS171 and ARG169 are both presents. The C-H ortho to the oxazole is making a long contact to HIS171 (3.6 Å, this could also be considered a side-on pi-pi contact) and the oxygen of the oxazole has contact with ARG169 at 3.5-3.6 Å. No other major contacts are present.
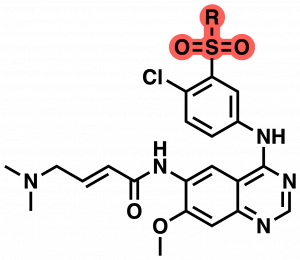
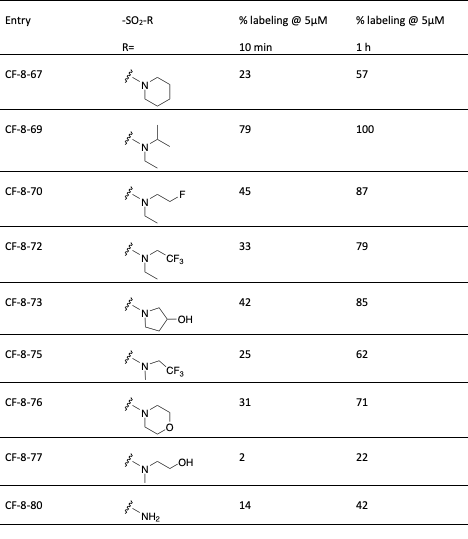
Table 1: SAR of CF-8-38 sulfonamide.
With this first round of compounds tested we continued to screen a variety of anilines but focused more heavily on modifications around the sulfonamide functional group as this had been the best compound in this first round assay. In the next round of compounds, new sulfonamide analogs were identified with increased labeling efficiency compared to CF-8-38 (59% labeling @5 µM over 1h) (Table 1). The greatest improvement was found by the replacement of one of the ethyl groups of the sulfonamide with an iPr group (CF-8-69). This gave 100% labeling over 1h and 79% labeling over 10 minutes @5µM. This is identical to improvements seen in our Benzamide series. From this data set, we can see that increasing size and hydrophobicity in general improve labeling in this assay. For example, this can be seen between CF-8-72 and CF-8-75 with an increase in labeling between the Me and Et groups, as well as the previously discussed improvement from CF-8-38 to CF-8-69 (Et to iPr). The free amine CF-8-80 also highlights the sensitivity in this region to changes in substituent size and hydrophobicity. The final interesting observation is the difference between CF-8-73 and CF-8-77. Both contain an exposed hydroxy group, but the ring-closed pyrrolidine gives a nearly 4-fold increase in labeling at 1 h and a 20-fold increase in 10 minutes. Finally, we examined methylation of the aniline at the 4-position of the quinazoline, this also gave improved mass spec labeling at 71% @ 5µM at the 1 h time point and 33% labeling at the 10 minute time point CF-8-78 (structure not shown).
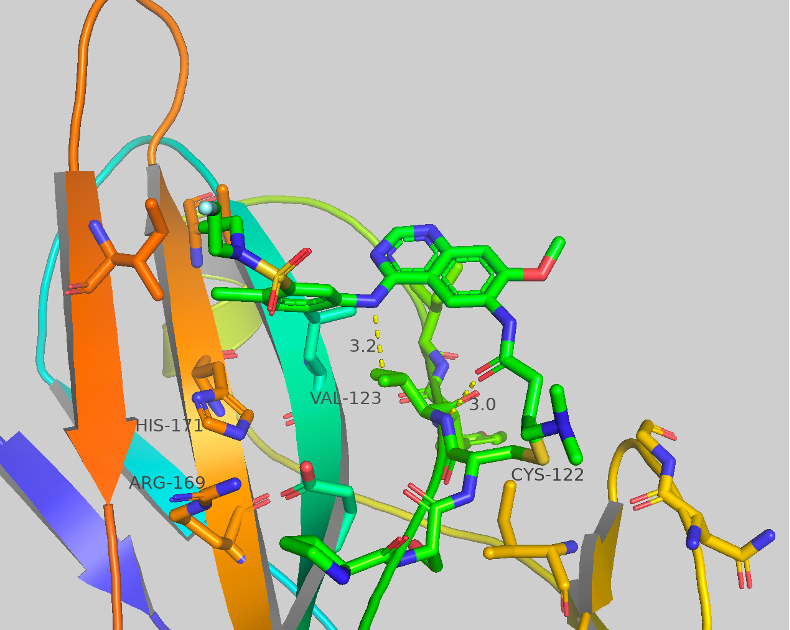
Figure 5 Crystal structure of CF-8-70 with contacts shown to corresponding amino acids.
We were able to generate crystal structures of some of the new compounds screened in the sulfonamide series. CF-8-70 is shown as an exemplar in Figure 5. Only two contacts are observed: to VAL-123 from the aniline NH (3.2 Å) and the carbonyl of the covalent warhead to a backbone NH (3.0 Å). No other major contacts are observed. Although many crystal structures we collect show little to no contact in the region of SAR modification, this highlights the high flexibility of this region as well as the potential for new structural changes to these compounds.
In conclusion, we have made meaningful progress on increasing the binding affinity (as measured by labeling efficiency) of compounds towards brachyury in two major series (Quinazoline and Benzamides). The best compounds have 8 to 10-fold increased labeling over 1h at 5 µM. We are now focused on increasing the labeling over 10 minutes and working towards 2 µM. Both series show further regions for diversification to continue making these improvements.
