I thought I’d spend a blog post just talking about the various paths of my research and try and tie it all together. A lot of my experiments do dot around these various arms of investigation and so one experimental post doesn’t necessarily tie in directly with the previous one or the next one and I thought it might be a good idea to explain why and summarise where I’m up to with each strand.
My main focus of research is to investigate the cause of the disease fibrodysplasia ossificans progressiva (FOP). We know that it’s caused by a mutation in ALK2 which leads to excessive bone formation. This also nicely dovetails with research done into DIPG where 25% of cases share a mutation with FOP in ALK2.
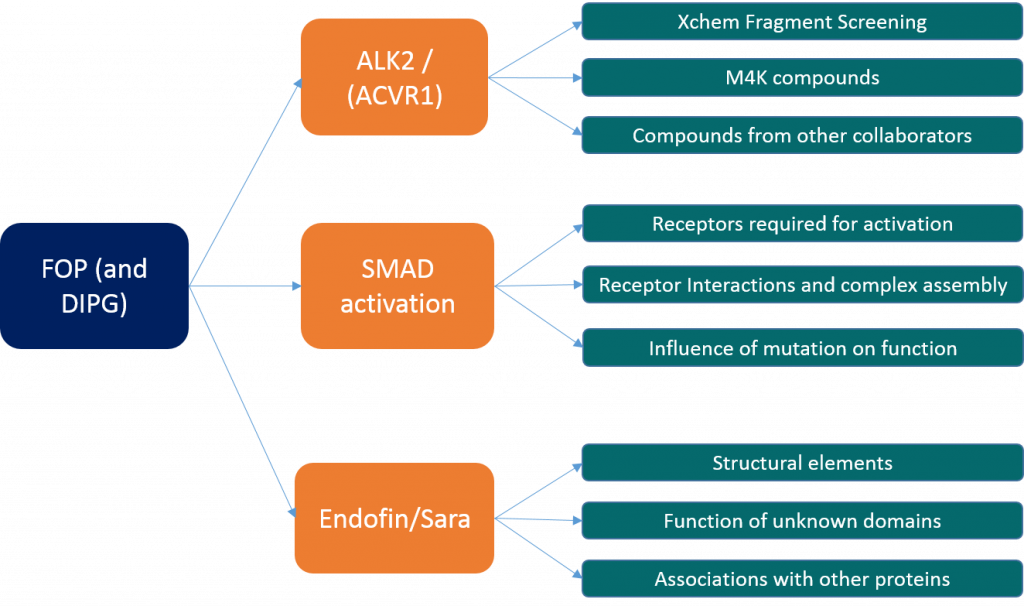
My research can be broken down in to three main strands:
- The first is looking at characterising the binding of ligands to ALK2 in order to optimise binding and work towards finding a potent and specific inhibitor. This is the focus of a lot of the M4K work that Jong Fu and Ros are doing and I’ve been doing a little bit of that as well but it’s not the main focus of my work. This endeavour is going fairly well – despite setbacks relating to protein expression (which are not entirely solved – we now have regained expression of the kinase domain alone but we still don’t have expression of the kinase domain and GS loop construct) and Ros has a number of structures that she’s working on at the moment that have been recently collected. Alas I’ve not had as much luck with new structures recently. We frequently get new deliveries of compounds from M4K to test – Jong Fu looks at them in cells while Ros and I try and crystallise the most promising candidates with varying degrees of success. Ros and I have also attempted XChem fragment screening which is where we use high throughput crystallography to identify small weakly bound fragments that might identify new leads for drug design or in our case, help us identify drugs that might bind to a second allosteric site within ALK2 away from the main ligand binding pocket. An XChem update will be coming soon from either Ros or myself as I’ve recently started helping her with this aspect of the work and so have been shadowing her on our last couple of visits to Diamond.
- The second is investigating the interactions of the type I receptor (ALK2) with its partnering type II in order to activate SMAD 1/5/8. This covers looking at receptor cross reactivity, SMAD cross reactivity, and the influence of mutations on this. This also involves looking at crystal structures of the mutants where possible, attempting to make complexes of the type I and type II receptors in order to try and crystallise them and probing the activities of the proteins and the various disease mutants. Covered in this is everything I’ve done using western blots and mass spec to identify the dependencies of this complex as required for SMAD phosphorylation. So far I know that the mutants are more active than the wild type (with R206H being the most active ALK2 mutation), that the type II must be present but does not need to be active to allow for SMAD activation and that increased activity is not solely dependent upon the removal of inhibition by FKBP12. Covered in this is also the work looking at mapping the phosphorylation sites in ALK2 to try and identify if some of these sites are more critical than others with relation to SMAD activation. The cloning I recently talked about is part of this investigation, making mutations to remove certain phosphorylation sites and comparing the activity of these with the wild type. As soon as these constructs have been cloned into suitable expression vectors I’ll be starting that experiment too. Another aspect of this is looking at the association of other proteins with the receptors, such as the possible link between them and XIAP as I talked about in one earlier blog post.
- The third aspect is looking other proteins related to the ALK2 pathway such as Endofin. Endofin is a membrane associated protein with multiple domains including a SMAD binding domain and large areas of unknown function. It is involved with the co-localisation of various parts of the pathway and appears to act as something of a scaffold protein to hold things in close proximity for signal propagation. The equivalent protein in the TGFβ pathway is called SARA and appears to have a similar function in that pathway. Some time ago I solved the structure for a domain of unknown function within SARA (4BKW) which contained a novel fold and showed structural (but not sequence) similarity to a protein called SUFU, which is part of the Hedgehog signalling pathway. We still don’t know what this protein and domain does or what it binds to or how it functions and so this is another area of investigation. If we can uncover the function of SARA we can also try and uncover the function of Endofin and this might provide a better understanding of the way FOP mutations work and help us see if there are any other ways we can target the pathway that are not exclusively based on inhibiting ALK2 activity. However given that the DUF domain has very little information about it, our investigations into what it does have been limited thus far. My goals with this are to try and probe the role of the DUF domain looking for whether it binds to either of the type I or type II receptors, and if so which parts of the receptors, or if it associates with a different protein all together. Given we know very little about what it does and the structure offers few clues (the lack of sequence similarity to SUFU means that although we can predict a possible binding mode, the nature of what binds there is very hard to determine as there are no conserved residues) this is quite a difficult problem to approach.
My work moves between these three strands depending on how much and which proteins I’ve got available to me – do I have any in the freezer already purified or do I need to make fresh? And that’s if I have the pellets in the freezer of the expressed protein in the first place. It also varies depending on whether we have new compounds we’re interested in testing immediately from either M4K or other collaborators, or whether I’ve got data I need to analyse from previous successful crystallisation attempts. Sometimes I’ll be looking at my own structural data and sometimes I’ll be looking at someone else’s structure which we call ‘proofreading’ to double check it for errors and make sure nothing has been missed. Sometimes we’ll have time at Diamond and so everything else has to be put on hold in order to make use of that visit (such as XChem trips) and sometimes there’s a lot of data analysis to do on other experiments – writing it up and understanding what the data is telling us is just as important for progress as the experiments themselves.
Having the three strands means I can move from one to another as each strand has its own peaks and lulls in activity at different times. I’m afraid it does make for a slightly more disjointed blog journey but I hope you’ll bear with me through it all as I try and make sense of the problems I’m investigating.
I’ll try and post again sooner rather than later on what I’ve been doing in January as it has been really rather bitty and frustrating in terms of things actually working but hopefully February will bring with it some more productive experiments.
