For clearer figures, please see this post at: https://zenodo.org/record/6993848#.Yv6Kwj3MI2w
Introduction
Project Proposal
The Structural Genomics Consortium (SGC) has recently embarked on its first research project in reproductive biology as a part of the SGC’s new open science Women’s and Children’s Health Program (WCHP). This program, which was generously funded by the Bill & Melinda Gates Foundation will focus on drug discovery for childhood diseases and development as well as reproductive biology and disease. One of the first initiatives of WCHP is to identify and characterize protein targets for non-hormonal contraceptive agents.
Throughout this article we will use the term people with uteruses (PWU) to encapsulate women, transgender men, and non-binary persons, all of whom experience unintended pregnancies. We will use female reproductive system to refer to the system, consisting of a uterus and ovaries, which can cause PWU to become pregnant. We note that the biological reproductive system may have no bearing on a person’s gender.
Contraception as Basic and Essential Healthcare
Access to contraception supports PWU’s autonomy over when and if to have children, which in turn has allowed PWU to pursue a career of their choosing, as well as financial stability. In 2005, Hock published research indicating that unrestricted access to contraceptives led to increased enrollment and graduation of PWU in college1. The ability to plan, delay, and space out births seems to empower PWU to achieve their educational and career goals2. Furthermore, delaying births can reduce the pay gap that exists between working PWUs and their peers who do not have children, as well as the gender pay gap2. Some disadvantages of unplanned births include: increased conflict and dissatisfaction in relationships, increased likelihood of experiencing anxiety and depression, obstacles to obtaining a secondary education, reduced earnings, and less preparation and financial resources to support the child2.
Birth control and contraceptive use not only supports PWU who want to avoid pregnancy; in many cases, birth control is prescribed as a method to treat another condition. Some examples include: to reduce pain in those with endometriosis, to protect against sexually transmitted infections such as pelvic inflammatory disease, for polycystic ovary syndrome, and to reduce hormonal acne. Birth control also impacts our society, economy, and planet. PWU who experience an unintended pregnancy are at a significant risk of mortality, morbidity, and financial hardship3. Additionally, our global population is increasing concurrent with an increase in average life expectancy, draining on our planet’s resources and stressing economies that attempt to support more people, for longer.
The Need for Effective Non-hormonal Contraceptives
Birth control enhances the lives of PWU and their partners, and advances our societies and economies; thus, it is imperative that PWUs have equal and uncomplicated access to birth control. As it stands, unintended pregnancy is a critical global issue, yet the rates of unintended pregnancy are higher among lower-income PWU and minorities3. Although access to contraception is an important factor to consider, other factors include the use of less efficient contraceptive methods, lack of patient education and understanding, as well as health concerns3. For example, according to a study published by the World Health Organization in 2020, approximately 40% of PWU using short-acting or long-acting modern contraceptive methods discontinued use due to side effects and health concerns4.
Currently, the most successful forms of birth control are permanent surgical procedures and hormonal methods. Unfortunately, these more effective birth control methods come at a price: extensive side effects. As most hormonal birth control methods target the female reproductive system, PWU are disproportionately affected by the negative side effects which range from moderate, such as dizziness, weight gain, painful cramping, diminished libido, and mood swings, to severe or even deadly, such as blood clots, high blood pressure, and depression. Due to the extensive side effects of hormonal birth control, it should be of the upmost priority to develop safer and more effective non-permanent contraceptive methods that align with PWU’s needs and preferences. In line with this, we aim to identify and test protein targets- in both male and female reproductive systems- as targets for non-hormonal birth control.
PATL2: A Potential Contraceptive Target
One of our selected protein targets, and the focus of this article, is Protein Associated with Topoisomerase II Homolog 2 (PATL2), an mRNA-binding protein that has also been shown to act as a repressor of translation5. PATL2 is expressed in immature oocytes and its expression decreases as oocytes mature6. PATL2 has been found to repress translation in oocytes where its temporal control is important for the maturation of oocytes: in the absence of PATL2 expression levels of certain genes involved in oocyte maturation and embryonic development were deregulated6,7. Furthermore, biallelic mutations in PATL2 led to reduced PATL2 protein levels causing fertilization failure and early embryonic arrest6. An excess of PATL2 can also be detrimental: in Xenopus oocytes, overexpression of PATL2 led to oocyte maturation arrest8. Overall, studies indicated that mutations or deregulation of PATL2 can cause female reproductive system infertility5,6,9.
PATL2 is a homolog of PATL1, which also contains a C-terminal PAT1 domain, and the two proteins share 38% identity. Both PATL1 and PATL2 are homologs of Pat1p in yeast5. In general, PAT proteins contain a conserved N-terminal sequence, a Mid domain, a proline-rich region, and a C-terminal domain5,10. Within the C-termini of the protein, a PAT1 domain (amino acids 252-491) is involved in PATL2’s mRNA-binding capabilities (Figure 1)9. As recently as 2020, 68% of disease-causing mutations identified in PATL2 were enriched in the PAT1 domain (aa 252-491), 32% were located in the N-terminal region (aa 1-250), with only one mutation identified in the C-terminus (aa 491-543)9. In contrast to the C-termini of PATL2, the N-terminal half of the protein (approximately 53% of the 543 amino acids) is predicted to be disordered (Figures 1 & 2). This is not altogether surprising, however, as intrinsically disordered regions (IDRs) are enriched in RNA-binding proteins relative to the entire human proteome11.
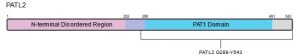
Figure 1: Schematic of full-length human PATL2 showing relevant domains.
Although there is very little information in the literature about the function of PATL2, there are some papers that provide information. To date, research suggests that PATL2 does not lead to global repression of RNA, but instead appears to regulate specific genes during meiosis II and the germinal vesicle stage that are critical for oocyte meiotic progression and early embryonic development7. Additionally, we can look to the more well-studied homolog PATL1 for insight. PATL1 has been shown to couple mRNA decapping and deadenylation in the 5’-3’ decay pathway through its interactions with the Lsm 1-7 complex12,13. PATL2 is capable of associating with some of the same proteins as PATL1, such as Lsm1 and Lsm4, of the Lsm1-7 complex, as well as Xrn113. As PATL2 has amino acid residues that correspond to the basic patch on PATL1’s C-termini that allows it to bind RNA, it is likely that PATL2 also retains similar RNA-binding properties5,14. Conversely, PATL2 exhibits some functions distinct from PATL1, and lacks others: unlike PATL1, PATL2 does not localize to P-bodies, and PATL1 does not compensate for PATL2’s role in meiosis5,13. Throughout our research, we will look to PATL1 as a springboard for our investigation into the function of PATL2, and its viability as a non-hormonal contraceptive target.
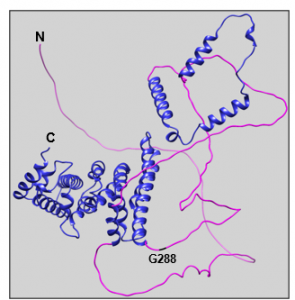
Figure 2: Predicted structure of full-length human PATL2 from AlphaFold2. The N and C-termini are labeled in black, along with amino acid G288, which serves as the start site for one of our PATL2 constructs. The disordered N-termini of the protein (aa 1-290) is colored magenta, while the ordered C-termini containing the PAT1 domain is colored in blue (aa 291-543).
Methods
Protein Production
To design PATL2 constructs for expression and purification, the protein sequence was first input in the AlphaFold2 program to analyze the predicted structure of PATL2. The predicted structure revealed a possible druggable pocket around residues F343, L346, Q349, A358, F361, I389, H396, and Q431. However, this prediction must be confirmed by determining the experimental structure of PATL2. Sequence similarity approaches, informed by AlphaFold2, were used to design the domain boundaries for protein expression constructs. Since protein constructs must be tested experimentally to determine if they will express well and be stable in a recombinant expression system, we initially designed nested constructs. We screened for constructs that express regions of interest: full-length, N-terminally truncated, and the PAT1 domain of PATL2, with hexahistidine tags for crystallization, and biotin tags to support antibody generation and assay development.
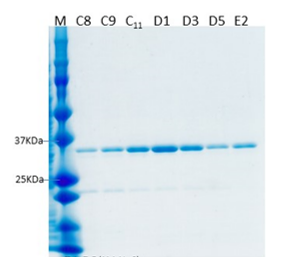
Expression constructs were first tested on a small scale to determine which constructs produced milligram quantities of soluble protein in E. coli or the SF9 insect cell/baculovirus system. From the constructs tested, we were able to successfully purify two PATL2 constructs from the SF9 insect cell/baculovirus system. The first was an N-terminally tagged hexahistidine protein construct consisting of amino acids G288-Y543, and the second consisted of the same amino acids from PATL2, but also contained a biotin tag. Both constructs were purified using cobalt bead affinity chromatography followed by gel filtration, and finally ion exchange chromatography (Figure 3).
In addition to these constructs, we are in the process of pursuing full-length human PATL2. As the first half of the protein is disordered, this creates challenges in purifying the full-length protein. The G288-Y543 PATL2 constructs lack this disordered region, likely part of the reason these constructs were able to be purified. It has previously been shown that maltose-binding protein (MBP) tags boost the expression and solubility of their fusion partners, and can even promote the proper folding of the attached protein into its biologically active conformation15,16. Moreover, it has been demonstrated that the MBP tag is successful at promoting the expression and solubility of other RNA-binding proteins with large disordered regions17. For these reasons, we are currently testing the expression of MBP-PATL2.
Figure 3: Purified PATL2 G288-Y543 with a biotin tag. The SDS-PAGE gel shows the purified protein elution samples from the final step of purification: ion exchange chromatography. The yield for this protein construct was several milligrams. Similar purity was obtained for the hexahistidine tagged PATL2 G288-Y543 construct (data not shown).
Structural Characterization
Since the PATL2 G288-Y543 constructs were stable and able to be purified, we set up several crystal trays to attempt to crystalize this compound. We have not yet observed crystals, but as the PAT1 domain of PATL1 has been crystallized, there is a high probability that we will be able to crystalize this domain in PATL2 as well10.
Functional Characterization
One of our first lines of investigation will be to determine if PATL2 can bind certain RNAs in vitro. As PATL1 has been found to bind G-quadruplexes, as well as poly(G) and poly(U) homopolymers, we will start by testing PATL2’s binding to these RNAs through fluorescence anisotropy, electrophoretic mobility shift assays, and competition assays. If we are able to obtain full-length PATL2, we will be able to compare it to PATL2 G288-Y543 to determine which regions of PATL2 contribute to its ability to bind RNA. If we confirm that PATL2 is capable of binding certain RNA sequences or structures, such as G-quadruplexes, we can then analyze the RNAs which PATL2 has been found to repress and determine if they contain these sequences/structures. Subsequently, we can test if PATL2 is able to bind in vitro to the RNAs it has been found to translationally repress in vivo.
Likewise, we would like to verify if PATL2 is able to directly bind proteins, such as Lsm1 and 4, to which it has been shown to associate (Figure 4)13,18. As an RNA-binding protein, it is likely that PATL2 is a part of an RNA-protein complex. Therefore, its interactions with other proteins may be mediated by RNA, as opposed to direct protein-protein interactions. For PATL1, it was shown that many of its interactions with other proteins were resistant to RNase A, indicating that these are protein-protein interactions that are not RNA-dependent13. It would be of interest to perform similar assays with PATL2 and its reported interactions partners Lsm1, Lsm4, and Xrn1. It is also a high priority for us to examine PATL2 interaction partners in oocytes. Again, if we can obtain full-length PATL2, we can compare it to PATL2 G288-Y543 to determine which regions of the protein contribute to its binding to its interaction partners.
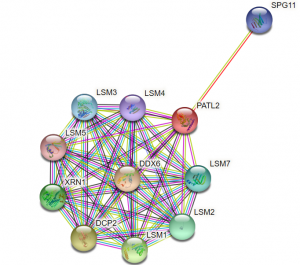
Figure 4: PATL2 protein-protein interactions from STRING. STRING is a functional protein association network that reports protein-protein interactions that are known based on experiments and curated databases, as well as predicted interactions (see reference 18). The interaction network provided for PATL2 will be used to guide our literature search into which PATL2 protein interaction partners to pursue for direct, in vitro binding validation.
Once we have developed a fuller understanding of the functions of PATL2, we will design assays to inhibit the protein’s RNA-binding or protein-binding function. As alluded to earlier, knowledge of which regions of the protein contribute to these functions will allow us to pinpoint regions of the protein to target for protein degradation or small molecule inhibitors. To assess small molecule inhibitors, we can employ assays such as thermal shift assays and isothermal titration calorimetry. Another approach to determining how to target PATL2 will be to analyze the PATL2 mutants that result in infertility. Several PATL2 mutations have been identified, and we can explore how they alter PATL2’s ability to bind RNA and to its protein interaction partners.
Acknowledgements
I would like to acknowledge Alma Seitova, Ashley Hutchinson, Maria Kutera, and Peter Loppnau for their contributions to clone and express PATL2 protein constructs. I would like to thank Hong Zeng for her work to purify the PATL2 constructs. I would also like to acknowledge Levon Halabelian and Aled Edwards for their input and supervision of this work.
This project is funded by the Bill and Melinda Gates Foundation. The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from AbbVie, Bayer AG, Boehringer Ingelheim, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], the EU and EFPIA through the Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and US), Pfizer, Takeda and the Wellcome Trust [106169/ZZ14/Z].
References
(1) Hock, H. The Pill and the College Attainment of American Women and Men. undefined 2007.
(2) Sonfield, A.; Hasstedt, K.; Kavanaugh, M. L.; Anderson, R. The Social and Economic Benefits of Women’s Ability To Determine Whether and When to Have Children. 48.
(3) Troutman, M.; Rafique, S.; Plowden, T. C. Are Higher Unintended Pregnancy Rates among Minorities a Result of Disparate Access to Contraception? Contracept. Reprod. Med. 2020, 5, 16. https://doi.org/10.1186/s40834-020-00118-5.
(4) Bellizzi, S.; Mannava, P.; Nagai, M.; Sobel, H. L. Reasons for Discontinuation of Contraception among Women with a Current Unintended Pregnancy in 36 Low and Middle-Income Countries. Contraception 2020, 101 (1), 26–33. https://doi.org/10.1016/j.contraception.2019.09.006.
(5) Maddirevula, S.; Coskun, S.; Alhassan, S.; Elnour, A.; Alsaif, H. S.; Ibrahim, N.; Abdulwahab, F.; Arold, S. T.; Alkuraya, F. S. Female Infertility Caused by Mutations in the Oocyte-Specific Translational Repressor PATL2. Am. J. Hum. Genet. 2017, 101 (4), 603–608. https://doi.org/10.1016/j.ajhg.2017.08.009.
(6) Chen, B.; Zhang, Z.; Sun, X.; Kuang, Y.; Mao, X.; Wang, X.; Yan, Z.; Li, B.; Xu, Y.; Yu, M.; Fu, J.; Mu, J.; Zhou, Z.; Li, Q.; Jin, L.; He, L.; Sang, Q.; Wang, L. Biallelic Mutations in PATL2 Cause Female Infertility Characterized by Oocyte Maturation Arrest. Am. J. Hum. Genet. 2017, 101 (4), 609–615. https://doi.org/10.1016/j.ajhg.2017.08.018.
(7) PATL2 Is a Key Actor of Oocyte Maturation Whose Invalidation Causes Infertility in Women and Mice. EMBO Mol. Med. 2018, 10 (5), e8515. https://doi.org/10.15252/emmm.201708515.
(8) Nakamura, Y.; Tanaka, K. J.; Miyauchi, M.; Huang, L.; Tsujimoto, M.; Matsumoto, K. Translational Repression by the Oocyte-Specific Protein P100 in Xenopus. Dev. Biol. 2010, 344 (1), 272–283. https://doi.org/10.1016/j.ydbio.2010.05.006.
(9) Liu, Z.; Zhu, L.; Wang, J.; Luo, G.; Xi, Q.; Zhou, X.; Li, Z.; Yang, X.; Duan, J.; Jin, L.; Zhang, X. Novel Homozygous Mutations in PATL2 Lead to Female Infertility with Oocyte Maturation Arrest. J. Assist. Reprod. Genet. 2020, 37 (4), 841–847. https://doi.org/10.1007/s10815-020-01698-6.
(10) Braun, J. E.; Tritschler, F.; Haas, G.; Igreja, C.; Truffault, V.; Weichenrieder, O.; Izaurralde, E. The C-Terminal Alpha-Alpha Superhelix of Pat Is Required for MRNA Decapping in Metazoa. EMBO J. 2010, 29 (14), 2368–2380. https://doi.org/10.1038/emboj.2010.124.
(11) Neelamraju, Y.; Hashemikhabir, S.; Janga, S. C. The Human RBPome: From Genes and Proteins to Human Disease. J. Proteomics 2015, 127, 61–70. https://doi.org/10.1016/j.jprot.2015.04.031.
(12) Sharif, H.; Conti, E. Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic MRNA Turnover. Cell Rep. 2013, 5 (2), 283–291. https://doi.org/10.1016/j.celrep.2013.10.004.
(13) Ozgur, S.; Chekulaeva, M.; Stoecklin, G. Human Pat1b Connects Deadenylation with MRNA Decapping and Controls the Assembly of Processing Bodies. Mol. Cell. Biol. 2010, 30 (17), 4308–4323. https://doi.org/10.1128/MCB.00429-10.
(14) Marnef, A.; Maldonado, M.; Bugaut, A.; Balasubramanian, S.; Kress, M.; Weil, D.; Standart, N. Distinct Functions of Maternal and Somatic Pat1 Protein Paralogs. RNA 2010, 16 (11), 2094–2107. https://doi.org/10.1261/rna.2295410.
(15) Costa, S.; Almeida, A.; Castro, A.; Domingues, L. Fusion Tags for Protein Solubility, Purification and Immunogenicity in Escherichia Coli: The Novel Fh8 System. Front. Microbiol. 2014, 5, 63. https://doi.org/10.3389/fmicb.2014.00063.
(16) Kapust, R. B.; Waugh, D. S. Escherichia Coli Maltose-Binding Protein Is Uncommonly Effective at Promoting the Solubility of Polypeptides to Which It Is Fused. Protein Sci. Publ. Protein Soc. 1999, 8 (8), 1668–1674. https://doi.org/10.1110/ps.8.8.1668.
(17) Edwards, M.; Xu, M.; Joseph, S. A Simple Procedure for Bacterial Expression and Purification of the Fragile X Protein Family. Sci. Rep. 2020, 10 (1), 15858. https://doi.org/10.1038/s41598-020-72984-7.
(18) Szklarczyk, D.; Gable, A. L.; Nastou, K. C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N. T.; Legeay, M.; Fang, T.; Bork, P.; Jensen, L. J.; von Mering, C. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49 (D1), D605–D612. https://doi.org/10.1093/nar/gkaa1074.
UniProt ID: C9JE40