The Background
In the scope of the SGC’s (Structural Genomics Consortium) Women’s and Children’s Health Initiative, a group of proteins that are potential targets for new, safe, and effective non-hormonal contraceptives will be characterized. These will then used as a template to screen for possible chemical probes and drug candidates. A group of postdoctoral researchers is funded by the Bill & Melinda Gates Foundation to tackle this challenge with each of them championing one specific target protein. I am one of these champions and want to introduce my target – ABHD2 – as well as present our results so far.
The α/β Hydrolase Domain 2 – ABHD2 – is part of the a/b Hydrolase Domain (ABHD) family, an in most parts poorly characterized group of hydrolases suggested to be involved in lipid metabolism, regulation, and signaling.1 Most of the 23 family members are predicted to have an active site consisting of a catalytic triad Ser-Asp-His. ABHD2 is a 48 kDa extracellular protein anchored to the plasma membrane via a single transmembrane helix.
Although found in multiple tissues, ABHD2 has been shown to play an essential part in sperm hyperactivation – the process that kicks the motility of a sperm into overdrive. The sperm faces several obstacles on its way to the oocyte, some of which can be overcome by this increase in motility, including viscoelastic luminal fluids, invasion of a cluster of cells around the oocyte – the cumulus oophorus – and eventually the zona pellucida, a layer of glycoproteins surrounding the oocyte.2 This hyperactivation is initiated by an influx of Ca2+ ions through the sperm-specific Ca2+-channel CatSper, located at the flagellum of the sperm. CatSper is inhibited by 2-arachidonoyl glycerol (2AG), an endocannabinoid of which substantial amounts are present in the sperm membrane. The degradation of 2AG necessary for hyperactivation is catalyzed by ABHD2, which is also located in the sperm flagellum. However, ABHD2 is in turn only activated in the presence of progesterone, which is secreted by cumulus cells.2,3 Inhibition of ABHD2’s hydrolase activity would therefore prevent opening of CatSper and subsequently hyperactivation of the sperm. In this context, testosterone, and the plant triterpenoids pristimerin and lupeol have been shown to avert sperm hyperactivation in the presence of progesterone, which might be promising starting points to identify chemical probes as well as drug candidates that inhibit ABHD2.4
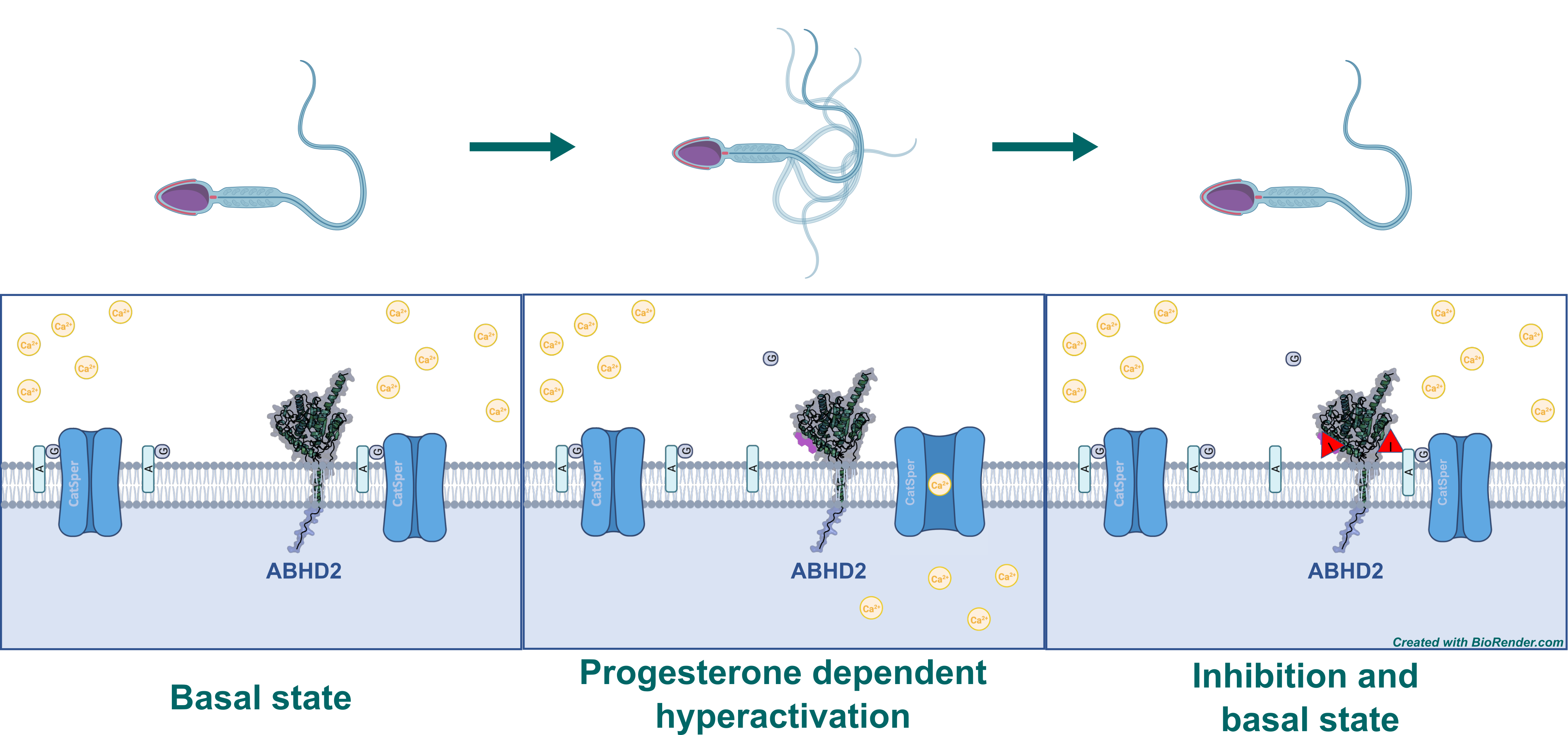
Figure 1: Mechanism of sperm hyperactivation by the degradation of 2AG mediated by ABHD2 upon binding of progesterone. 2AG gets digested to glycerol (G) and arachidonic acid (A). Inhibitor of ABHD2 is indicated by “I” in red triangle.
The Goal
We aim to generate a Target Enabling Package – TEP – for ABHD2. This includes the production, structural and biophysical characterization, assay development, and the generation of recombinant antibodies, but also screening for chemical probes and of course drug candidates to facilitate the development of non-hormonal contraceptives.
Additionally, other ABHD proteins will be produced to analyze potential off-target effects of chemical probes and drug precursors.
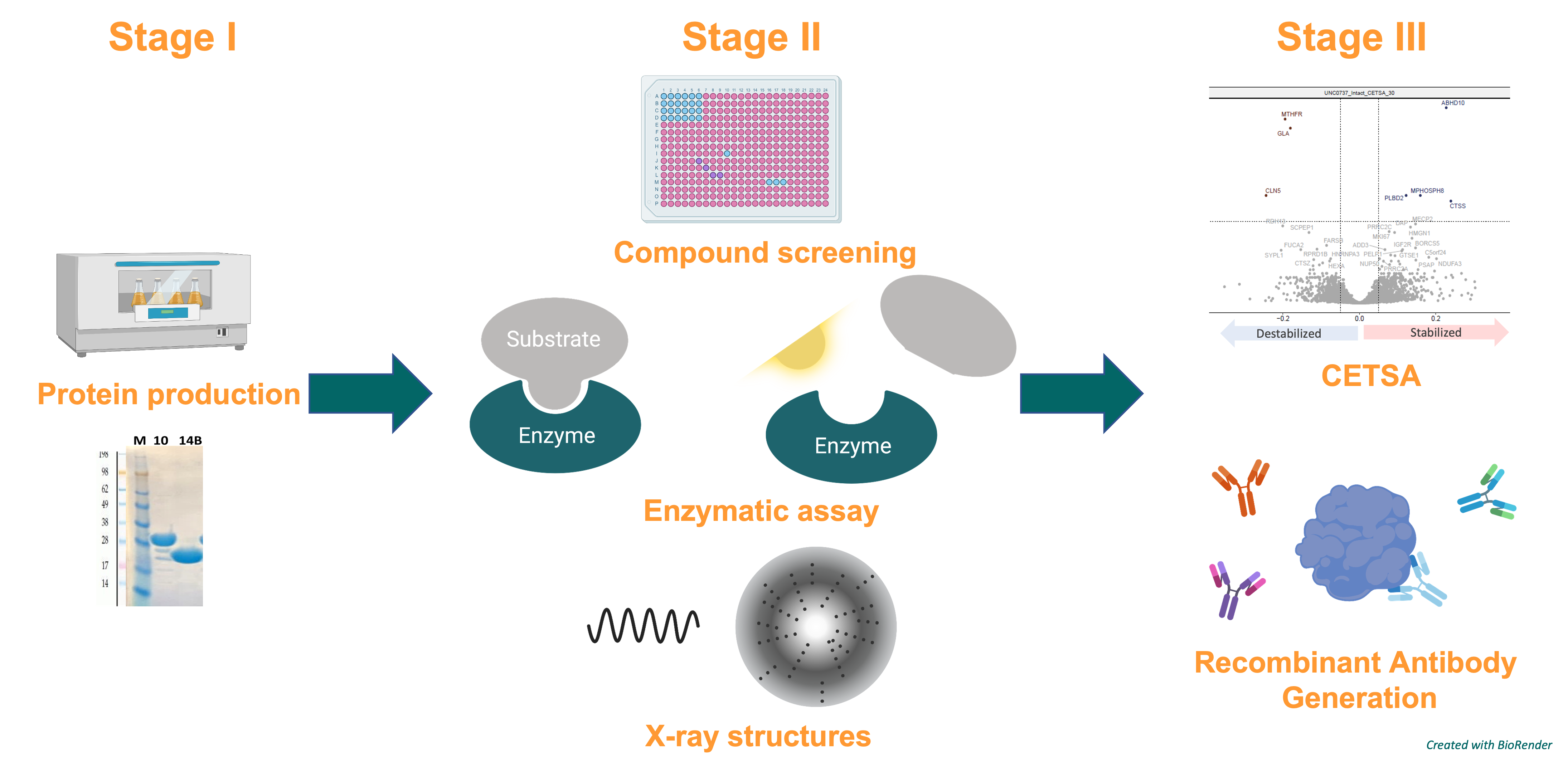
Figure 2: General procedure for generating a TEP for ABHD2.
The Results
Production of ABHD2 in E. coli
To produce the protein, a diverse set of constructs was designed with varying lengths to screen for fragments that are soluble and suited for crystallization (see Table 1).5,6 The constructs were based on the sequence-based prediction of a transmembrane helix (L10-V30) and the position of the core a/b-hydrolase domain (M128-G382), as well as positions of terminal secondary elements based on structure prediction by AlphaFold (ID: AF-P08910-F1).
ABHD2 fragments were inserted in pNIC28-Bsa4 and pNIC-NT6HB vectors via the LiC-cloning method.7 Both vectors carry a N-terminal hexa-His-tag followed by a recognition sequence for TEV-protease for cleaving, while the fragments in pNIC-NT6HB are additionally fused with a sequence for biotinylation after the N-terminal His-tag. The latter is required for downstream generation of recombinant antibodies. Also, both vectors carry in their original form the SacB gene, which is to be replaced by the target fragment. In the presence of 5% sucrose serves as a suicide gene, i.e., lysing cells that contain this particular gene. This allows for negative selection, allowing only cells that carry vectors with desired target fragment to survive on plates with sucrose. Insertion of ABHD2-fragments was further verified by colony-PCR.
Table 1: Cloned constructs for E. coli expression
| Construct-ID | Label | Vector | Residues | MW (Da) |
| ABHD2-c1_Bsa4 | 1 | pNIC28-Bsa4 | N2-G382 | 45842.95 |
| ABHD2-c2_Bsa4 | 2 | pNIC28-Bsa4 | N2-E425 | 50867.62 |
| ABHD2-c3_Bsa4 | 3 | pNIC28-Bsa4 | R31-G382 | 42846.35 |
| ABHD2-c4_Bsa4 | 4 | pNIC28-Bsa4 | R31-E425 | 47871.02 |
| ABHD2-c5_Bsa4 | 5 | pNIC28-Bsa4 | D43-G382 | 41567.82 |
| ABHD2-c6_Bsa4 | 6 | pNIC28-Bsa4 | D43-E425 | 46592.48 |
| ABHD2-c7_Bsa4 | 7 | pNIC28-Bsa4 | K74-G382 | 37971.57 |
| ABHD2-c8_Bsa4 | 8 | pNIC28-Bsa4 | K74-E425 | 42996.24 |
| ABHD2-c9_Bsa4 | 9 | pNIC28-Bsa4 | H93-G382 | 35903.15 |
| ABHD2-c10_Bsa4 | 10 | pNIC28-Bsa4 | H93-E425 | 40927.81 |
| ABHD2-c11_Bsa4 | 11 | pNIC28-Bsa4 | R31-A403 | 45251.16 |
| ABHD2-c12_Bsa4 | 12 | pNIC28-Bsa4 | D43-A403 | 43972.62 |
| ABHD2-c13_Bsa4 | 13 | pNIC28-Bsa4 | K74-A403 | 40376.38 |
| ABHD2-c14_Bsa4 | 14 | pNIC28-Bsa4 | H93-A403 | 38307.95 |
| ABHD2-c15_Bsa4 | 15 | pNIC28-Bsa4 | G96-G382 | 35505.71 |
| ABHD2-c16_Bsa4 | 16 | pNIC28-Bsa4 | G96-A403 | 37910.52 |
| ABHD2-c17_Bsa4 | 17 | pNIC28-Bsa4 | G96-E425 | 40530.38 |
| ABHD2-c18_Bsa4 | 18 | pNIC28-Bsa4 | V129-G382 | 31880.65 |
| ABHD2-c19_Bsa4 | 19 | pNIC28-Bsa4 | V129-A403 | 34285.46 |
| ABHD2-c20_Bsa4 | 20 | pNIC28-Bsa4 | V129-E425 | 36905.32 |
| ABHD2-c4_NT6HB | Bio4 | pNIC-NT6HB | R31-E425 | 47871.02 |
| ABHD2-c6_NT6HB | Bio6 | pNIC-NT6HB | D43-E425 | 46592.48 |
| ABHD2-c8_NT6HB | Bio8 | pNIC-NT6HB | K74-E425 | 42996.24 |
| ABHD2-c10_NT6HB | Bio10 | pNIC-NT6HB | H93-E425 | 40927.81 |
With the final constructs at hand, we were ready to start initial expression screens in small scale. This was done according to the SGC standard protocol in BL21(DE3)-R3-pRARE2 cells transformed with the various constructs. They were incubated in 1.4 ml Terrific Broth (TB) medium supplemented with 50 µg/ml kanamycin and 34 µg/ml chloramphenicol 37 °C until an OD600nm (Optical Density) of 2 – 3. After cooling to 18 °C, expression of the fragments was induced by adding 0.5 mM IPTG and incubating overnight (ca. 20 h).
To distinguish between soluble and insoluble proteins, samples of the harvested cells after chemical lysis for the insoluble (total) fractions were taken. The rest of the lysate was cleared by centrifugation and incubated with Ni2+-Sepharose Fast Flow beads at 4 °C for 1,5 h. As all fragments of ABHD2 were fused with an N-terminal hexa-His-tag, soluble ABHD2 should coordinate to complexed Ni2+-cations on the Sepharose beads. After washing, immobilized proteins were eluted and collected. Samples from this contained the soluble protein. Both fractions for every construct were analyzed by SDS-PAGE. Coomassie stained gels are displayed in Figure 3. Labels are as shown in Table 1 and correspond to respective constructs.
While all constructs showed high expression levels, only for ABHD2-c4_Bsa4, ABHD2-c10_Bsa4, and ABHD2-c17_Bsa4 small amounts of soluble ABHD2-fragments were observed, indicated by orange arrows (Figure 3). Despite a high background of unspecifically bound proteins, these three constructs were subsequently tested in up scaled expression.
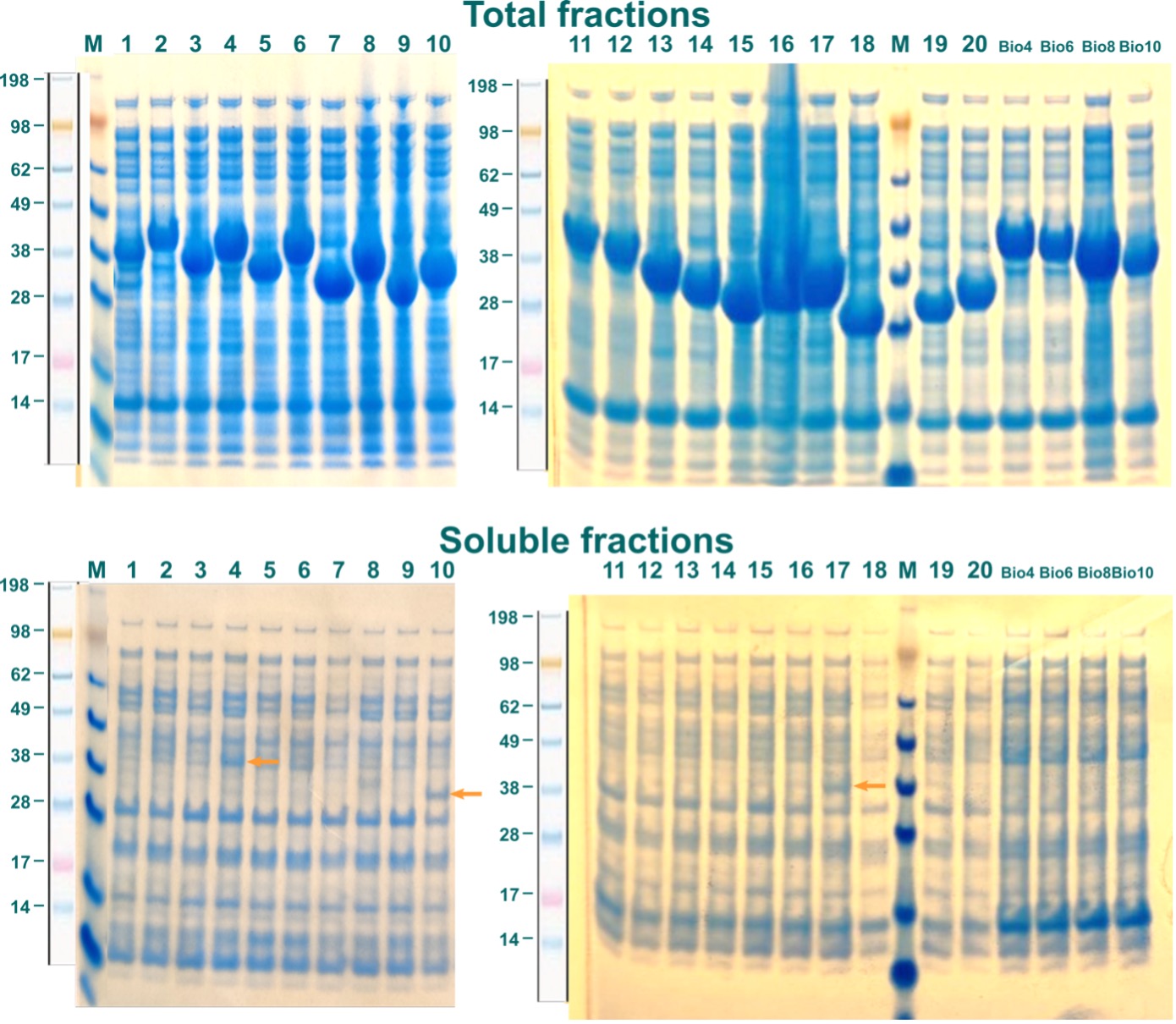
Figure 3: Expression of ABHD2 constructs in E.coli. Constructs are indicated by labels as in Table 1. Soluble ABHD2-fragments are indicated by orange arrows.
For large-scale cultures 0.75 l TB medium were inoculated from 20 ml LB (Lysogeny-Broth) starting culture, which was incubated overnight and started in turn from a glycerol stock of the particular constructs transformed in BL21(DE3)-R3-pRARE2 cells. As described above, media were supplemented with 50 and 34 µg/ml of kanamycin and chloramphenicol, respectively. Cultures were incubated analogously as in the expression screen. After harvesting, re-suspended cell pellets were frozen at -80 °C and thawed before being lysed by sonication and cleared by centrifugation.
Purification of the three constructs was approached with a 2-step process, consisting first of an IMAC (Immobilized Metal Affinity Chromatography) followed by a SEC (Size Exclusion Chromatography). In this scheme, proteins are specifically coordinated first to the solid phase of the column by binding a complexed (immobilized) bivalent metal-cation, here Ni2+, via the hexa-His-tag. After washing, bound protein was eluted in one step and subsequently separated by size using SEC. Elution profiles of this last step for the three ABHD2-fragments are illustrated in Figure 4. Colored arrows in the chromatograms indicate fractions analyzed by SDS-PAGE, labelled correspondingly in the same color above the respective gel. Orange arrow in the gel denotes position of individual ABHD2 fragment.
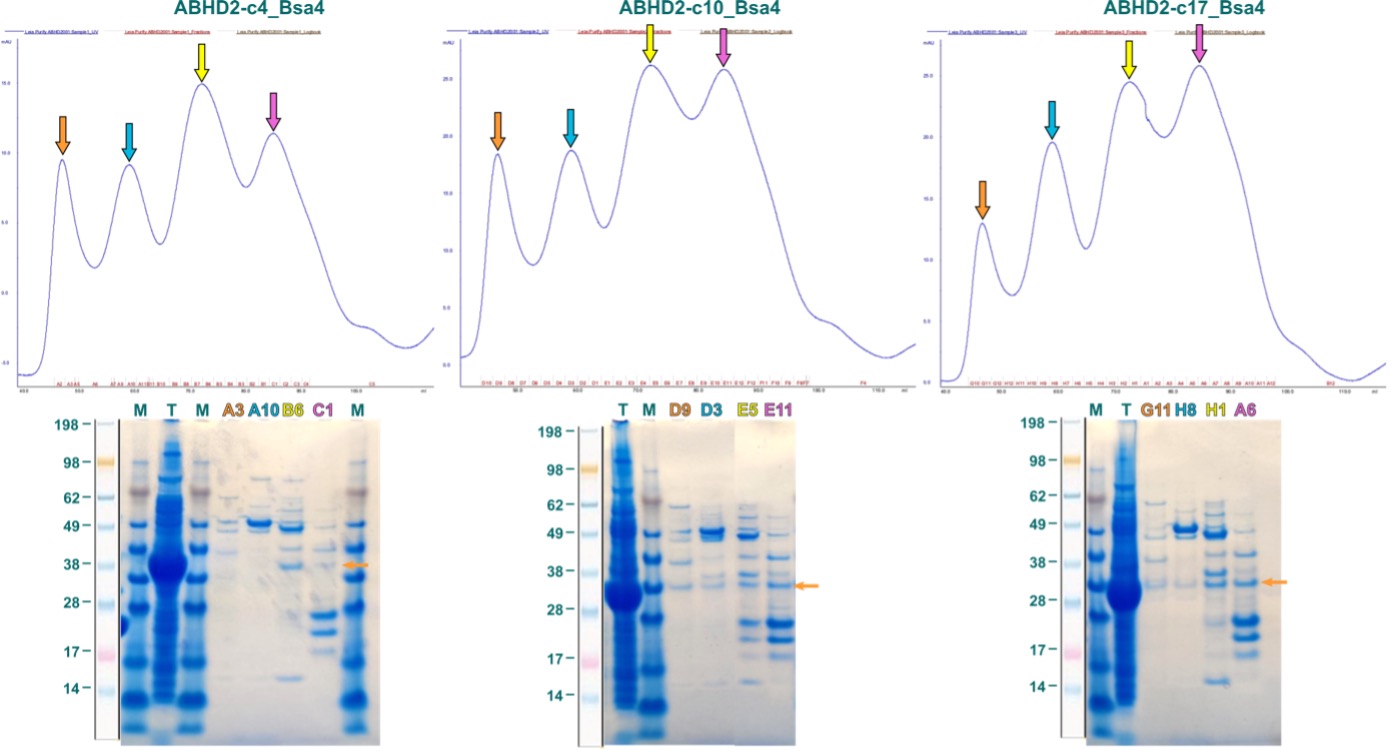
Figure 4: First purification attempts for ABHD2 fragments expressed in E. coli. Displayed are SEC elution profiles above with the respective SDS-PAGE below. Colored fraction labels correspond to same-colored arrows in elution profile. Orange arrow in the gel indicates position of ABHD2 fragment based on band from total (T) lysate.
As shown in Figure 4, the elution profiles of all three ABHD2 fragments, for which small amount of soluble protein was observed in the expression screening, are ambiguous with four absorption A280nm maxima. Fractions corresponding to these peaks comprised multiple bands, however, bands that agree with the molecular weight of the ABHD2 fragment and are also observed for the total lysate (T) were very weak.
Expressing ABHD2 in E. coli proved to be quite challenging. Reasons for this can be manifold – the protein being extracellular, membrane-anchored, and having 14 cysteine residues are only first possible explanations that come to mind. Therefore, we’ve moved to the Baculovirus Expression Vector System (BEVS), which is better suited for more challenging proteins.
Production of ABHD2 in Baculovirus Expression Vector System
As for E. coli, several constructs of ABHD2 were created consisting of different combinations of His-, Flag, and Avi-tags, displayed in Table 2.
Table 2: Constructs cloned for expression BEVS.
| Construct-ID | Label | Vector | Residues | MW (Da) |
| ABHD2_37-425_BV_CHisFlag | D10 | pBacMam2-DiEx-LIC | S37-E425 | 47881.76 |
| ABHD2_37-425_BV_CHis | D11 | pFB-CHis-LIC | S37-E425 | 45616.41 |
| ABHD2_37-425_BVs_AviN-CHis | D12 | pFHMSP-AviN-LIC-C | S37-E425 | 51140.73 |
| ABHD2_37-425_BVs_Nhis-AviC | E01 | pFHMSP-AviC-LIC-N | S37-E425 | 52454.22 |
| ABHD2_37-425_BV_Nhis | E02 | pFBOH-MHL | S37-E425 | 46624.53 |
| ABHD2_37-425_BV_AviN-Chis | E03 | pFBD-BirA | S37-E425 | 48014.96 |
| ABHD2_37-410_BV_CHisFlag | E04 | pBacMam2-DiEx-LIC | S37-N410 | 46191.94 |
| ABHD2_37-410_BV_CHis | E05 | pFB-CHis-LIC | S37-N410 | 43926.59 |
| ABHD2_37-410_BVs_AviN-CHis | E06 | pFHMSP-AviN-LIC-C | S37-N410 | 49450.91 |
| ABHD2_37-410_BVs_Nhis-AviC | E07 | pFHMSP-AviC-LIC-N | S37-N410 | 50764.4 |
| ABHD2_37-410_BV_Nhis | E08 | pFBOH-MHL | S37-N410 | 44934.71 |
| ABHD2_37-410_BV_AviN-Chis | E09 | pFBD-BirA | S37-N410 | 46325.14 |
| ABHD2_91-425_BV_CHis | E11 | pFB-CHis-LIC | S91-E425 | 39585.3 |
| ABHD2_91-425_BVs_AviN-CHis | E12 | pFHMSP-AviN-LIC-C | S91-E425 | 45109.62 |
| ABHD2_91-425_BVs_Nhis-AviC | F01 | pFHMSP-AviC-LIC-N | S91-E425 | 46423.11 |
| ABHD2_91-425_BV_Nhis | F02 | pFBOH-MHL | S91-E425 | 40593.41 |
| ABHD2_91-425_BV_AviN-Chis | F03 | pFBD-BirA | S91-E425 | 41983.84 |
These SF9 cells were transfected with these constructs and screened for expression. Like screenings in E. coli, cells were lysed and soluble fractions were incubated with Ni2+-beads. After washing, beads were incubated with elution buffer and the resulting eluates analyzed via SDS-PAGE (Figure 5).
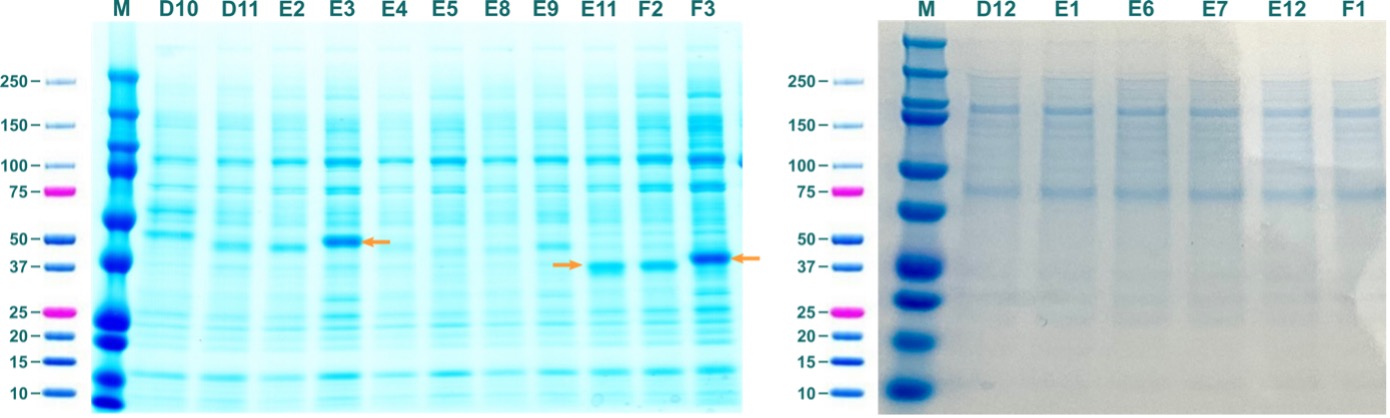
Figure 5: Soluble fractions from test expressions of ABHD2 constructs in BEVS. Labels of lanes correspond to labels of constructs in Table 2. Orange arrows indicate prominent bands that correspond to molecular weight of respective ABHD2 constructs.
SDS-PAGE revealed prominent bands that correspond to the molecular weight of soluble ABHD2 for ABHD2_37-425_BV_AviN-Chis (E03; 48014.96 Da), ABHD2_91-425_BV_CHis (E11; 39585.3 Da), and ABHD2_91-425_BV_AviN-Chis (F03; 41983.84 Da).
Until now constructs “E3” and “E11” were tested in scaled-up expression, but unfortunately initial results suggest that a major part of ABHD2 is expressed in insoluble inclusion bodies or soluble aggregates. However, there are still a lot of conditions to be tested and we are optimistic to get soluble ABHD2 for further downstream assays!
Off-Targets
When developing inhibitory compounds, specificity is one of the major concerns one must consider. For this reason, not only ABHD2 but also other members of the ABHD family will be produced and analyzed for off-target effects of potential chemical probes and drug candidates.
The process of creating constructs and expression screenings for other ABHDs was analogously to what is described above for ABHD2. From the tested constructs only fragments for ABHD10, ABDH11, and ABHD14B showed soluble expression. Out of those, the longest variants without signal sequences were chosen for further purification, corresponding to ABDH10-c1_Bsa4 (T53-N306; 30887.54 Da), ABHD11-c2_Bsa4 (V34-V315; 33600.47 Da), and ABHD14B-c1_Bsa4 (A2-Q210; 24898.40 Da).
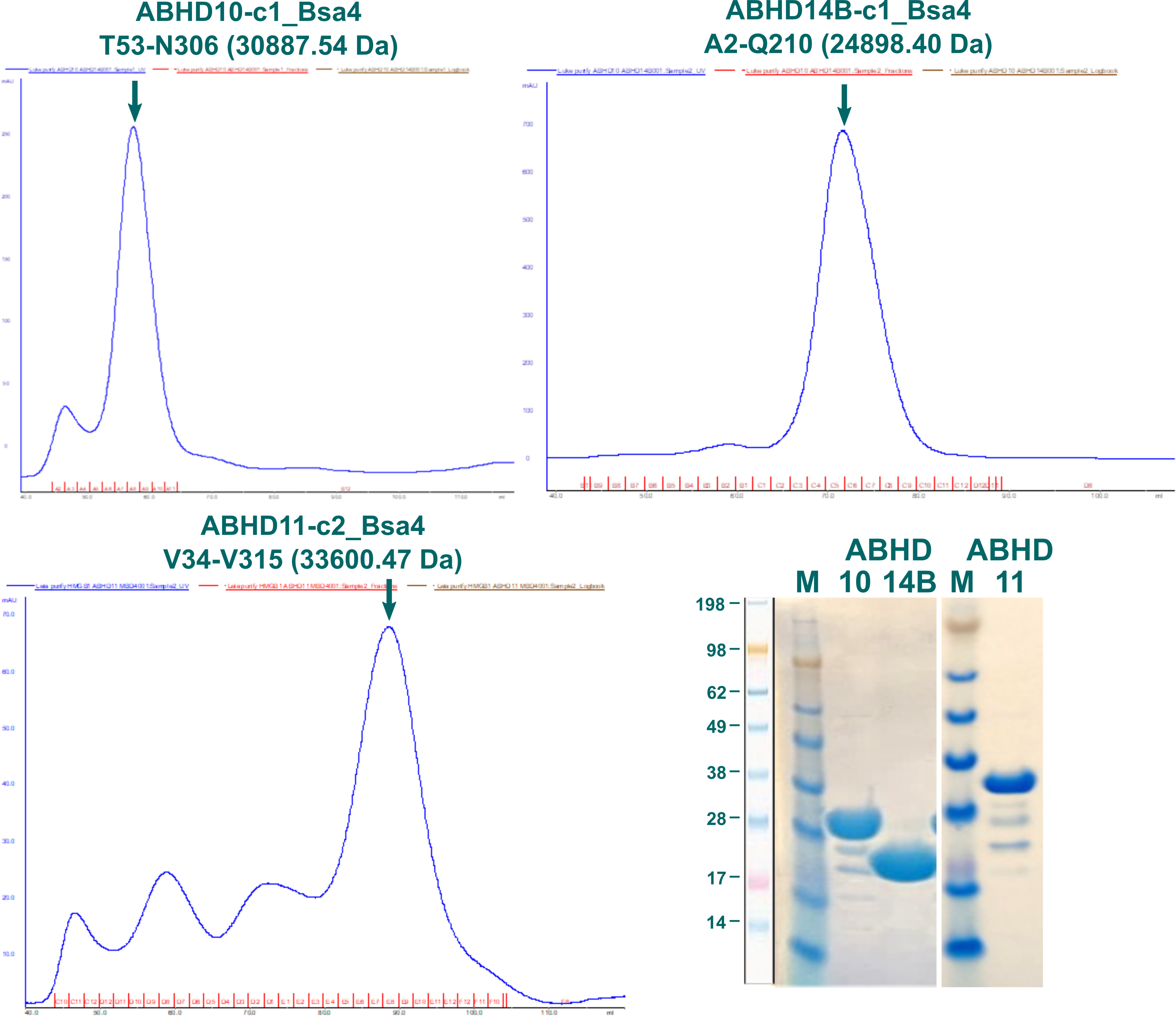
Figure 6: SEC elution profiles of ABHD10, ABHD11, and ABHD14B fragments alongside SDS-PAGE of pooled fractions.
Elution profiles for three fragments of ABHD10, ABHD11, and ABHD14B are displayed in Figure 6 alongside SDS-PAGE of pooled fractions indicated by a green arrow. Proteins were stored at -80 °C in 20 mM HEPES, 300 mM NaCl, 5 % glycerol, 0.5 mM TCEP, pH7.5.
To test activity of purified fragments and quantify degree of inhibition of compounds, an enzymatic assay had to be developed. Protocols for these kind of activity assays are already described in the literature for some ABHDs, most of which used p-nitrophenyl derivatives as substrates. 8–10 In these assays, the p-nitrophenyl substrate is hydrolyzed, forming p-nitrophenol in the process whose absorption can be colorimetrically detected at 405 nm. The protocols had to be optimized for a 96-well plate format to ensure a time-efficient procedure with low amounts of protein and compound consumption.
Testing different incubation times and temperatures as well as different substrate and protein concentrations yielded conditions, under which the increase in product absorbance still shows linear behavior and consuming less than 10 % of substrate but with a clearly detectable signal above noise levels. These conditions are important to ensure a steady-state system in which concentration of the substrate is in excess to the enzyme. This is necessary to assume a constant concentration of the enzyme-substrate complex, a prerequisite for Michaelis-Menten kinetics.
Conditions were found for ABHD10 and ABHD11 that used 300 and 10 nM enzyme respectively, 500 µM p-nitrophenyl butyrate, in 20 mM HEPES, 150 mM NaCl, pH7.5, 2 % DMSO, 1.4 % Methanol with incubation in 200 µl at 37 °C for 20 min. Figure 7 shows the linear increase in absorbance over time for ABHD10 and ABHD11 at different substrate concentrations. Data was recorded in triplicates and absorbance was referenced to identical preparations without protein at every time point. DMSO was added to directly test conditions at which inhibitory compounds can be tested and methanol was used as solvent for the substrate.
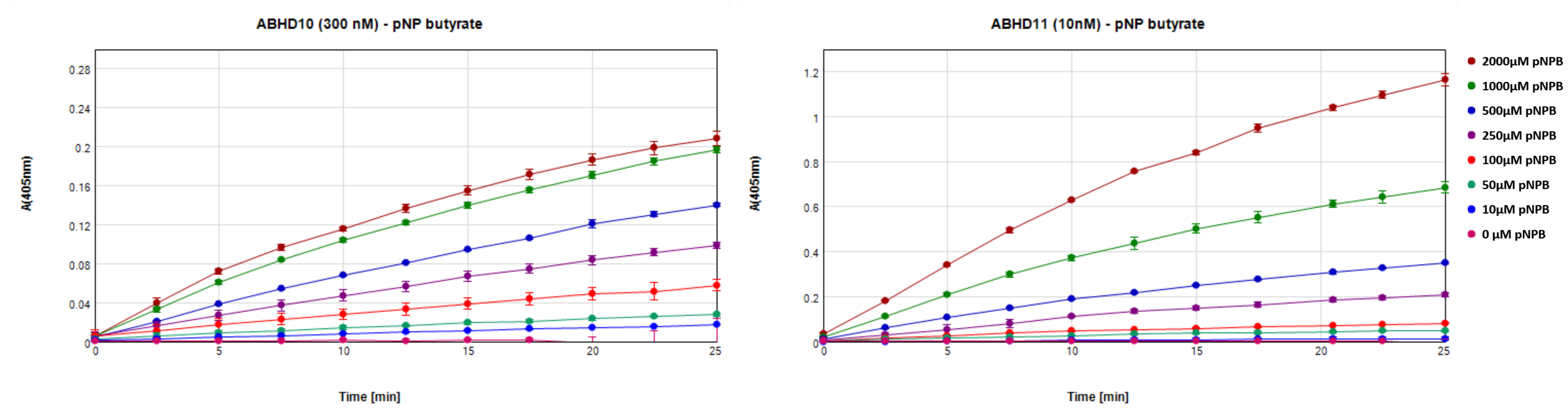
Figure 7: Increase in absorbance at 405 nm over time for incubation of 500 µM p-nitrophenyl butyrate with 300 nM ABDH10 or 10 nM ABHD11.
As indicated in the literature, p-nitrophenyl substrates appear to be degraded by most ABHDs, including ABHD2, which suggests that the conditions found for ABHD10 and ABHD11 can be swiftly adapted for ABHD2.
Another critical aspect of our endeavor is to identify highly specific chemical probes and drug candidates. Identifying suitable compounds for not just ABHD2 but other ABHDs can not only aid in understanding biological function but especially help creating specific compounds by investigating off-target effects. Also, identifying promiscuous compounds might facilitate the generation of a specific compound. Even minute differences can create or abolish a specific interaction and, in fact, is one of the criteria for the SGC’s catalogue of chemical probes.11
A serendipitous result in an unrelated project found a compound that bound very selectively to ABHD10 in a Cellular Thermal Shift Assay (CETSA), UNC0737 (Figure 8). Due to the quinazoline core of the compound we screened a library of 264 quinazoline compounds provided by Peter Brown from the University of North Carolina at Chapel Hill in a Differential Scanning Fluorimetry (DSF) assay for interactions with ABHD10. DSF is a variation of a Thermal Shift Assay (TSA) that measures the changes of fluorescence along an increasing temperature gradient. Proteins lose their native structure at higher temperatures and unfold. During this, hydrophobic sidechains that would normally form the hydrophobic core of the polypeptide get exposed to the solvent. SYPROTM orange, a fluorescent dye added beforehand to the sample mixture but whose fluorescence is quenched in water, then binds to these exposed hydrophobic sidechains. Because of this interaction the fluorescence of the dye is no longer quenched and a fluorescence signal can be detected (excitation at 492 nm; emission at 610 nm). The sigmoidal change in fluorescence dependent on the temperature can be fitted and the inflection point of the curve is defined as the melting point – Tm – of the protein. Upon binding a compound, a protein is stabilized in most cases, as the complex is thermodynamically favorable compared to both single molecules (the protein and the compound). This will lead to an increased melting point for the complex compared to the free protein, as more energy, i.e., higher temperature, is required to denature the stabilized form.
With this we identified nine compounds from the library that increased the melting point compared to free ABHD10 by more than 2.5 °C (Figure 8).
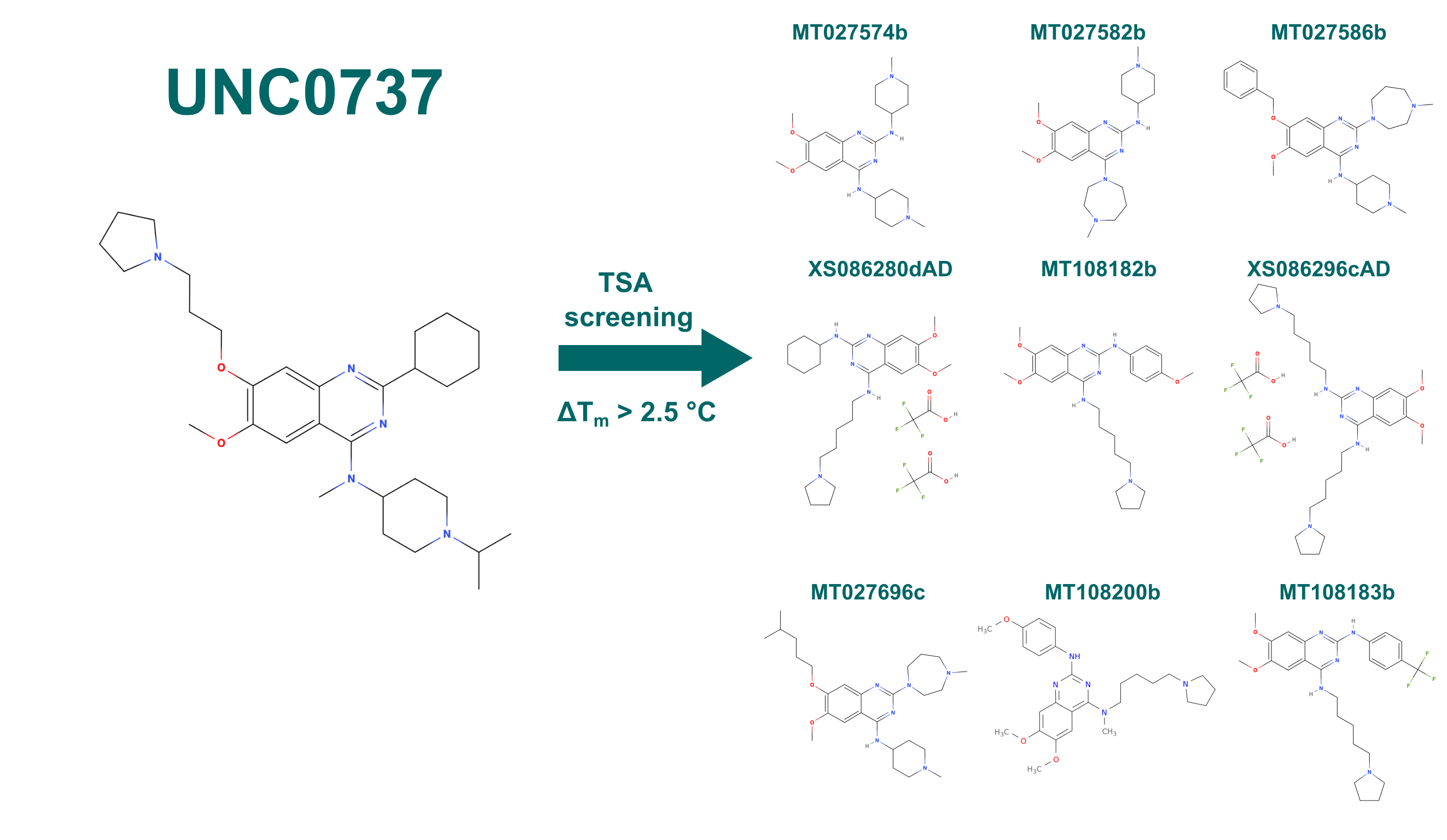
Figure 8: Compounds that stabilized ABHD10 by more than 2.5 °C in a DSF screen. Library was based on the quinazoline core structure of UNC0737.
The Outlook
Expanding on the promising results from test expression we will screen more conditions for expression and purification of ABHD2 in BEVS on a larger scale. Further, constructs for secreted proteins will be tested as well as potentially cell-free expression approaches to tackle the challenges of expressing ABHD2 from multiple angles. The produced protein will be checked for activity in an enzymatic assay using p-nitrophenyl derivatives as substrates and similar incubation conditions that were already established for ABHD10 and ABHD11. Further, inhibitory compounds of ABDH2 will be screened for using DSF as well as an inhibition enzymatic assay to quantify the potency of the compounds. As there is no experimental structure available, crystallization trials for the free and complexed protein will be initiated, as well, to gain a better understanding of ABHD2 on a structural level and identify interaction sites.
You can also find a version of this post on Zenodo.
Acknowledgement
I would like to thank Alma Seitova, Madison Edwards, and Levon Halabelian for their excellent help with the expression of ABHD2 in BEVS and Edvard Wigren for his help with the protein production from E. coli and showing me around. Also, Evert Homan and Elisee Wiita for helping with screening of compounds as well as Peter Brown for providing the compound library.
References
- Lord, C. C., Thomas, G. & Brown, J. M. Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochimica et Biophysica Acta – Molecular and Cell Biology of Lipids vol. 1831 792–802 Preprint at https://doi.org/10.1016/j.bbalip.2013.01.002 (2013).
- Trebichalská, Z. & Holubcová, Z. Perfect date—the review of current research into molecular bases of mammalian fertilization. Journal of Assisted Reproduction and Genetics vol. 37 243–256 Preprint at https://doi.org/10.1007/s10815-019-01679-4 (2020).
- Miller, M. R. et al. Unconventional endocannabinoid signaling governs sperm activation via the sex hormone progesterone. Science (1979) 352, 555–559 (2016).
- Mannowetz, N., Miller, M. R. & Lishko, P. v. Regulation of the sperm calcium channel CatSper by endogenous steroids and plant triterpenoids. Proc Natl Acad Sci U S A 114, 5743–5748 (2017).
- Gräslund, S. et al. The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expression and Purification 58, 210–221 (2008).
- Structural Genomics Consortium et al. Protein production and purification. 5, (2008).
- Strain-Damerell, C., Mahajan, P., Gileadi, O. & Burgess-Brown, N. A. Medium-Throughput Production of Recombinant Human Proteins: Ligation-Independent Cloning. in Structural Genomics: General Applications (ed. Chen, Y. W.) 55–72 (Humana Press, 2014). doi:10.1007/978-1-62703-691-7_4.
- Rajendran, A., Vaidya, K., Mendoza, J., Bridwell-Rabb, J. & Kamat, S. S. Functional Annotation of ABHD14B, an Orphan Serine Hydrolase Enzyme. Biochemistry 59, 183–196 (2020).
- Bailey, P. S. J. et al. ABHD11 maintains 2-oxoglutarate metabolism by preserving functional lipoylation of the 2-oxoglutarate dehydrogenase complex. Nature Communications 11, (2020).
- Naresh Kumar, M. et al. Molecular characterization of human ABHD2 as TAG lipase and ester hydrolase. Bioscience Reports 36, (2016).
- Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nature Chemical Biology vol. 11 536–541 Preprint at https://doi.org/10.1038/nchembio.1867 (2015).
