Special thanks to:
David Drewry – Helped with designing the nanoBRET tracers
M4K pharma OICR chemist team – Synthesised the M4K1046-linker derivatives
Carrow Wells (UNC) – Conjugated the M4K1046 to nanoBRET fluorophore
Background:
I have always wanted to establish nanoBRET target engagement assay for ALK5. Relative to dual luciferase promoter assay and immunofluorescent staining, nanoBRET is many times faster. With a robust ALK5 nanoBRET assay, I will be able to rapidly screen for cellular off-target activity.
Since none of the commercial nanoBRET tracers worked with ALK5, we attempted to generate our own tracers. We chose to create these bespoke tracers based on M4K1046 because it has cellular IC50 of ~50nM for ALK5. David Drewry has helped to design linkers that will attach fluorophores (nanoBRET energy acceptor) to the solvent-facing end of M4K1046. Based on known structures, additional bulk in this region should not hinder the binding of the compounds to ALK5. Two versions of M4K1046 (with different linker length) were synthesised by the M4K pharma chemist team in OICR. They were subsequently sent to Carrow Wells for conjugation to nanoBRET fluorophore.
Experimental design:
I wanted to determine the binding of these tracers to ALK5-nanoluciferase fusion. Therefore, I incubated increasing concentrations of tracer with HEK293 cells expressing ALK5-nanoluciferase fusion protein. If the tracers can bind nicely to the ATP pocket of ALK5, incubation with increasing concentrations of tracer will result in increasing BRET signal (wavelength = 610 nm). The resulting magnitude of BRET and EC50 estimated from the curve are good indicators to whether these tracers can be used for nanoBRET assay with ALK5.
To identify any background signal, I have included a replicate of the above experiment with 10,000nM of parent M4K1046. These unlabelled compound will saturate ALK5 ATP pockets and prevent the binding of nanoBRET tracers. All signal from this set of experiment is contributed by non-specific background.
To be eliminate doubts of any technical or reagent issues with the nanoBRET experiment itself, I have replicated both experiments above, substituting ALK5 with ALK2.
Results:
EGFP signal in transfected cells. HEK293 were transfected efficiently. Cells can be harvested for nanoBRET assay.
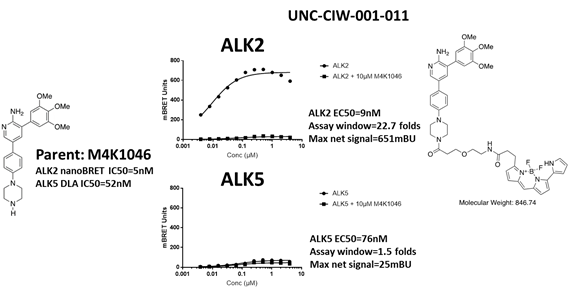
EC50 curves of the first bespoke ALK5 tracer (UNC-CIW-001-011) with ALK2-nanoluc and ALK5-nanoluc. The chemical structures of parent M4K1046 and the bespoke tracer are shown on the side of the graphs. This tracer cannot bind to ALK5.
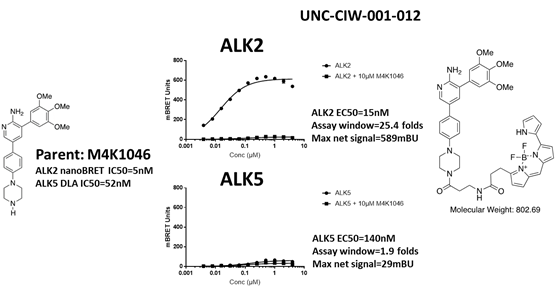
EC50 curves of the second bespoke ALK5 tracer (UNC-CIW-001-012) with ALK2-nanoluc and ALK5-nanoluc. The chemical structures of parent M4K1046 and the bespoke tracer are shown on the side of the graphs. This tracer cannot bind to ALK5.
Conclusion:
Both bespoke tracers cannot be used for ALK5 nanoBRET. Both of them did not bind to ALK5. The nanoBRET experiment itself was a success since both tracers worked well with ALK2 (over 20 folds assay windows). The background noise was low for both tracers. ALK5 seems to intrinsically not tolerate bulky additions on the solvent end of M4K1046. This might be additional avenue for improving ALK2 vs ALK5 selectivity. At the very least, we have gained this additional information from these efforts.
For additional experimental details, please refer to my Zenodo post.

Jong Fu, the tracers turn-out useless for ALK5, but any merit in using them for ALK2 NanoBRET? (And great to see the unexpected SAR insight to make ALK2 inhibitors even more selective!)
Dear Matthieu,
Thank you for the comment!
These custom-made tracers worked decently with ALK2 but not better than the current commercial Promega tracer. On top of that, more than 300 compounds has already been profiled using the commercial tracer.
Instead, when I get the time, I am intending to test them out for other Type I and Type II in the TGF-beta/BMP family since many of them still lack nanoBRET target engagement tracer. (fingers crossed)
It is really intriguing, structurally, what is causing ALK5 to not tolerate the additional bulk.
Hi Jong,
Did you ever contact Promega R and D to get any advice on tracer design? If its still something you are interested in I can always schedule meeting with Craig who’s the Project Manager for this product. He’s in touch with most of the labs doing NanoBRET TE work in the UK and he might be able to come up with some tips(he has done for my other customers in the past).
Regards,
David
Dear Jong,
Are you still looking for at ALK5 Tracer? If so are you in contact with Promega R and D? I know we are making new tracers all the time and screening them against the whole Kinome so there are far more tracers available than commercially advertised. I’ve asked if we have anything and I’ll let you know. I can also put you in touch with the product manager for NanoBRET TE if you want any tips on synthesising tracers if you want to try it again.
Regards
David
Dear David,
Thank you so much for the help!
We are indeed in touch with Matt and Jim from the Promega team. They had been tremendously helpful with our project!
Gratefully
Jong Fu