In my last post, I showed how we found the residues lining the catalytic site of SARS-CoV-2 SAM-dependent m7GpppA-specific 2’-O-methyltransferase (2’-O-MTase), which I refer to as NSP16.
In this post, I will share with the diversity dendrograms corresponding to the reviewed entries of the Alpha- and Betacoronavirus genera from the UniProt database. I use a 3D structure of NSP16 (PDB: 6wvn) to highlight the non-conserved residues across those sequences.
To look at the genetic variability of entries belonging to the Alpha- and Betacoronavirus genera, we used their reviewed sequences from UniProt. This time, our focus was on the 39 residues lining the NSP16 catalytic site. Here is a full list of these 39 residues: K6822, C6823, D6824, L6825, Y6828, N6841, K6844, Y6845, H6867, F6868, G6869, A6870, G6871, S6872, P6878, G6879, S6896, D6897, L6898, N6899, D6912, C6913, D6928, M6929, Y6930, D6931, P6932, T6934, K6935, V6937, F6947, K6968, T6970, E6971, H6972, N6996, S6999, S7000, E7001.
Altogether, there were 28 unique entries in UniProt for both Alpha- and Betacoronavirus genera that we aligned using multiple sequence alignment in ICM. I show The diversity dendrogram in Figure 1. The dendrogram highlights the diversity profile at each of the 39 positions and groups the entries from different organisms into separate clusters based on their sequence similarity. You can see in figure 1 that the entry SARS2 is highlighted in a red box and sits close to entry CVHSA, which is SARS-CoV organism, and they both are from the Betacoronavirus genus.
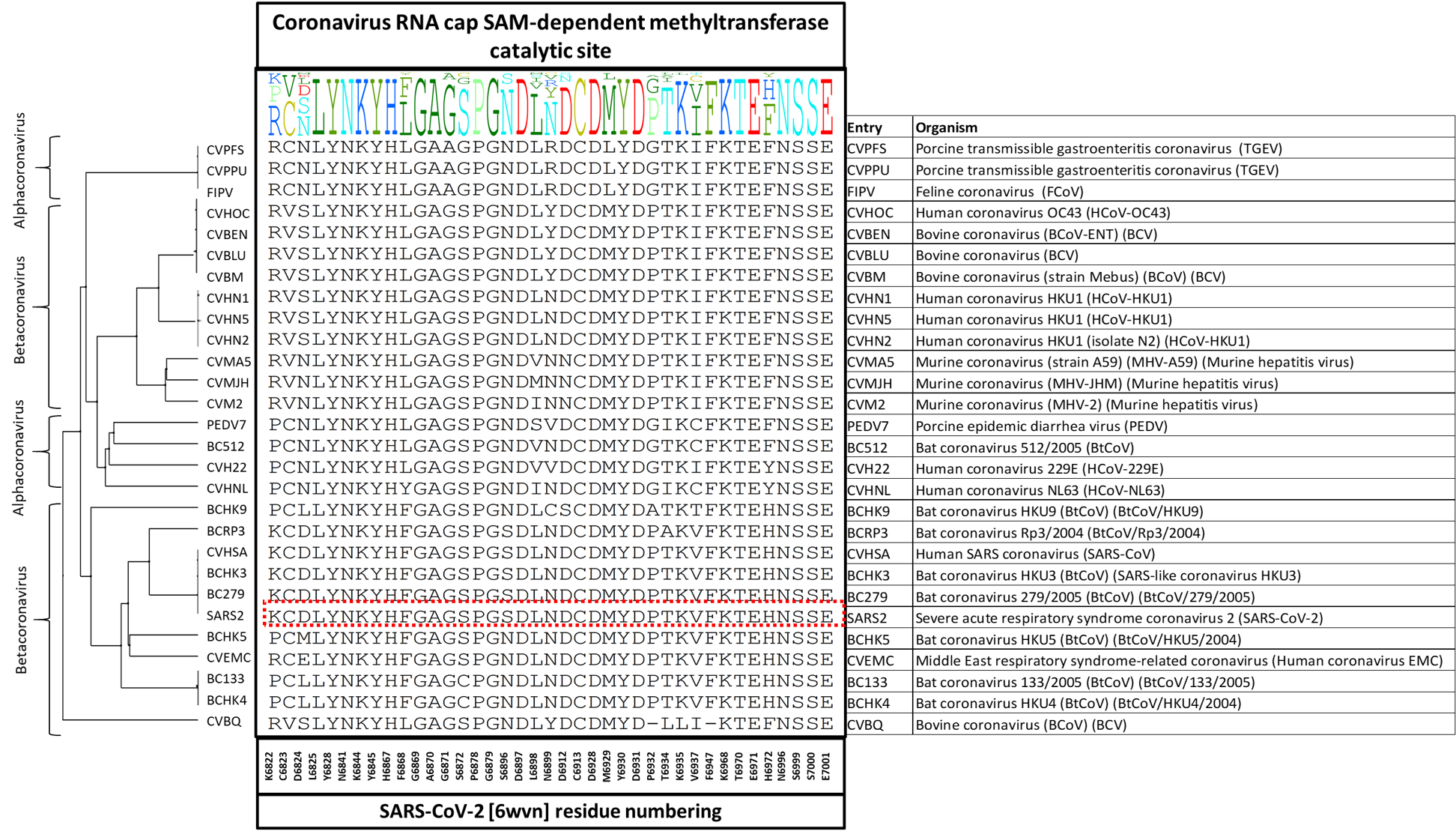
Figure 1. The diversity dendrogram of the catalytic site of 2’-O-MTase for the Uniprot entries of Alpha- and Betacoronavirus genera.
Figure 2 is the color-coded representation of SARS-CoV-2 2’-O-MTase to highlight the non-conserved residues of the Alpha and Betacoronavirus genera based on the diversity dendrogram in figure 1.
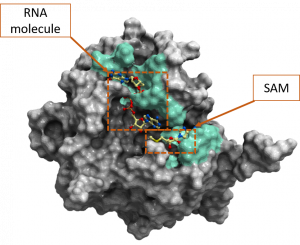
Figure 2. Color-coded representation of SARS-CoV-2 2’-O-MTase (PDB: 6wvn). The green color shows the non-conserved residues among the UniProt entries from the Alpha- and Betacoronavirus genera. These residues include K6822, C6823, D6824, F6868, G6871, S6872, S6896, L6898, N6899, D6912, M6929, P6932, T6934, K6935, V6937, H6972. Both the cofactor SAM and the RNA molecule are labeled on this figure.
Next, we looked at the conservation of 2’-O-MTase catalytic site residues among the 21 entries of the Betacoronavirus genus separately which is depicted in the diversity dendrogram in figure 3. The color-coded representation of SARS-CoV-2 2’-O-MTase in figure 4 highlights the non-conserved residues at NSP16 catalytic site among the entries in the Betacoronavirus genus.
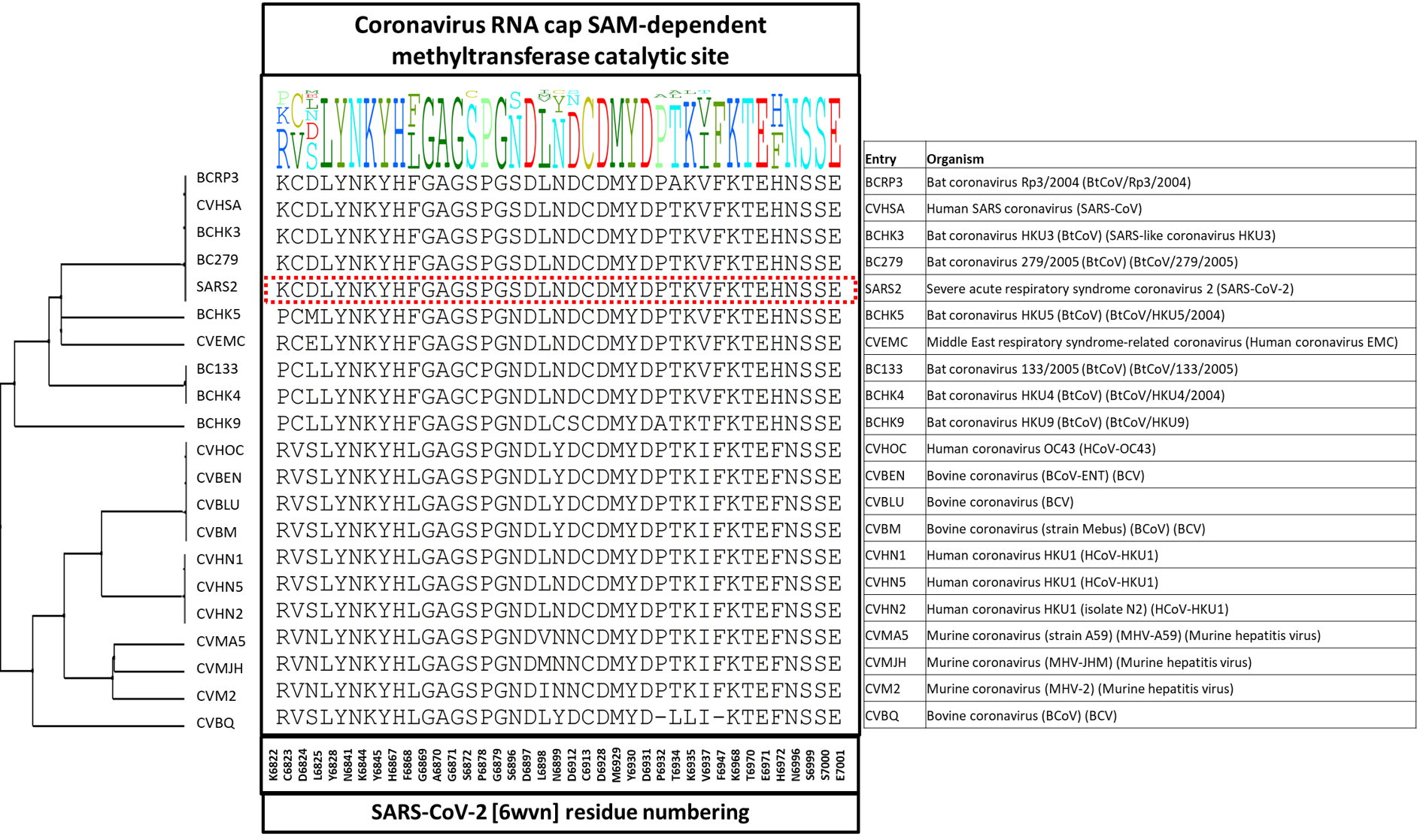
Figure 3. The diversity dendrogram of 2’-O-MTase catalytic site residues among the UniProt entries of the Betacoronavirus genus.
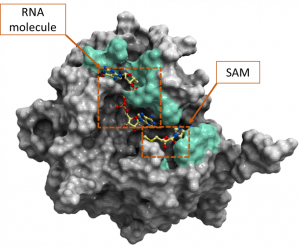
Figure 4. Color-coded representation of SARS-CoV-2 2’-O-MTase (PDB: 6wvn). The green color shows the non-conserved residues among the UniProt entries from the Betacoronavirus. These residues include K6822, C6823, D6824, F6868, S6872, S6896, L6898, N6899, D6912, P6932, T6934, K6935, V6937, H6972. Both the cofactor SAM and the RNA molecule are labeled on this figure.
To view the detailed steps, please see my Zenodo report here.
In my next post, I will show the energy calculations corresponding to the variants from SARS-CoV-2 patient samples at the catalytic site of NSP16.
As always, I would be happy to hear your comments. Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project.
