Within a month (hopefully) I’ll be convincing C2C12 myoblast cells to express all kinds of mutant ALK2 to test its activity and interactions with other proteins within the cell. I’ll be doing this by treating them so that they’ll take up pieces of circular DNA (plasmids) that express those mutant proteins (i.e. transfecting them).
Seeing as I’m going to be doing that countless times, it makes sense to find the most efficient way to do that, whilst I wait for extra plasmids I need to arrive. I’m looking for the way that will use the smallest amount of DNA and reagents, but will still transfect a high proportion of cells. So far, I’ve been using Lipofectamine-2000, which traps the DNA in a lipid capsule and helps the cell absorb it, but I heard that the chemical polyethylenimine (a positive compound that helps DNA to stick to the negative surface of the cell) could work just as well with less DNA.
To compare the two methods, I performed them both on the cells with a plasmid that will express enhanced green fluorescent protein (EGFP), because it visibly marks the cells which have been transfected:
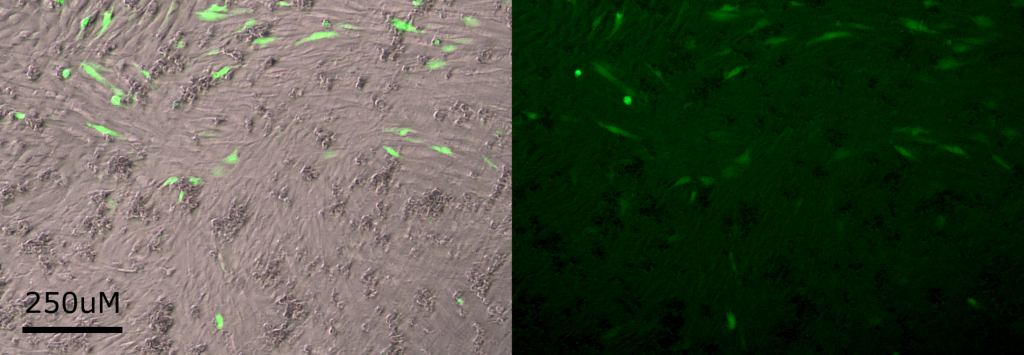
Left: A brightfield image of C2C12 cells overlaid with the fluorescent image from expressed EGFP.
Right: The fluorescent image alone.
Now I know that treating cells with PEI at a ratio of 4x the volume of PEI to the mass of DNA transfects more cells than the previous method that relied on Lipofectamine-2000. Full experimental details can be read here.

Elizabeth, do you have an electroporator? That works with about 50% efficiency, we did Crispr cas9 transduction that way.
Actually I saw someone in the department using one recently. Thanks for the idea I’ll look into the technique with them !