Background
PRMT6 is type I protein arginine methyltransferase which mono- and asymmetrically dimethylates histone and nonhistone proteins, mainly in glycine and arginine rich motifs (Fig.1). PRMT6 shows predominant nuclear localization and is implicated in the regulation of nuclear processes such as DNA repair and gene expression (PIMID: 11724789, 18079182).
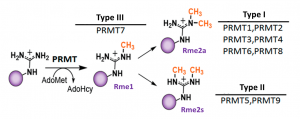
Fig.1. Mammalian PRMTs. There are 9 members and 3 types of the PRMT family. Type I, II and III PRMTs catalyze the formation of monomethyl arginines (Rme1). Type I PRMTs catalyze asymmetric arginine methylation (Rme2a), while type II PRMTs form symmetric arginine methylation (Rme2s). Type III PRMT can only monomethylate arginine residues. Modified from (PMID:29378138).
Assay validation
It has been reported that PRMT6 is the main contributor to histone H3 arginine 2 asymmetric dimethylation (H3R2me2a) in cells (PIMID: 17898714). We found that knocking down of PRMT6 for 3 days is not sufficient to observe a decent decrease in H3R2me2a levels, however, overexpression of wild type PRMT6 but not its catalytic mutant (V86K/D88A) led to increase in H3R2me2a levels (Fig.2). Overexpression of PRMT6 also increased H3 arginine 8 asymmetric dimethylation (H3R8me2a) and H4 arginine 3 asymmetric dimethylation (H4R3me2a) levels (Fig.2). The assay was further validated with PRMT Type I chemical probe (MS023). MS023 decreased PRMT6 dependent H3R2 asymmetric dimethylation in cells overexpressed with PRMT6 in a dose-dependent manner (Fig.3). The Z factor for the assay equals 0.63.
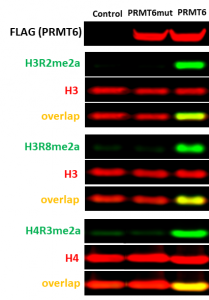
Fig.2. The wild type but not catalytic mutant of PRMT6 asymmetrically dimethylated endogenous H3R2, H3R8, and H4R3. HEK293T cells were transfected with FLAG-tagged PRMT6 wild type or V86K/D88A catalytic mutant (PRMT6mut) or empty vector (control) for 24 h. The methylation levels were analyzed in Western Blot.

Fig.3. MS023 decreases PRMT6 dependent H3R2 asymmetric dimethylation in cells. HEK293T cells were transfected with FLAG-tagged PRMT6 wild type or V86K/D88A catalytic mutant (PRMT6mut) and treated with inhibitor for 20 h. H3R2me2a levels were determined by Western blot. The graph represents the nonlinear fit of H3R2me2a signal intensities normalized to total histone H3. The results are mean +/- SEM of 3 replicates.
Experimental details are posted on Zenodo.
