Background
PRMT7 is a type III protein arginine methyltransferase that monomethylates arginine residues mainly within the RXR motif (Fig.1) (PIMID: 24247247). PRMT7 was shown to play an important role in muscle physiology, stem cell biology, and cellular stress response (PIMID: 32409666, 27225728, 26854227).
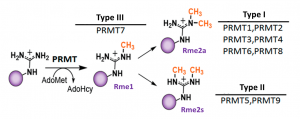
Fig.1. Mammalian PRMTs. There are 9 members and 3 types of the PRMT family. Type I, II, and III PRMTs catalyze the formation of monomethyl arginines (Rme1). Type I PRMTs catalyze asymmetric arginine methylation (Rme2a), while type II PRMTs form symmetric arginine methylation (Rme2s). Type III PRMT can only monomethylate arginine residues. Modified from (PMID:29378138).
Assay validation
It has been reported previously that PRMT7 is the main contributor to HSP70 R469 monomethylation in cells (PIMID:32409666). We found that PRMT7 knockout or knockdown reduces HSP70 monomethylation in various cell lines (Fig. 2). The assay was further validated with the PRMT7 selective chemical probe SGC3027 (PIMID: 32409666). SGC3027 but not its inactive control SGC3027N decreased PRMT7 dependent HSP70 monomethylation in C2C12 cells in a dose-dependent manner (Fig.3). The Z factor for the assay equals 0.67.
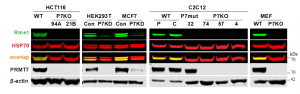
Fig. 2. PRMT7 (P7) knockout (KO) or knockdown (KD) reduces HSP70 methylation in various cell lines. Western blot analysis of HSP70 monomethylation. 94A, 21B—HCT116 CRISPR PRMT7 KO clones; P parental C2C12; C-C2C12 expressing control guide RNA; 32 C2C12 CRISPR clone expressing PRMT7 catalytic mutant (delY35,A35S); 4,57,74—C2C12 CRISPR Prmt7 KO clones. PRMT7 was knocked down in HEK293T and MCF7 cells using siRNA. Con-control, P7KD-PRMT7 knockdown. Modified from PIMID:32409666. (Shawna Organ, Magdalena Szewczyk)
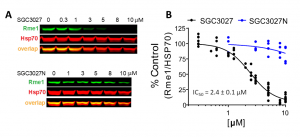
Fig.3. SGC3027 but not negative control compound SGC3027N inhibits PRMT7-dependent HSP70 monomethylation in C2C12 cells in a dose-dependent manner. Cells were treated with the compound for 2 days. A. A representative blot for SGC3027 and SGC3027N effects on HSP70 methylation. Rme1—arginine monomethylation.B Quantification of SGC3027 and SGC3027N effects on HSP70 monomethylation in C2C12 cells. The graphs represent non-linear fits of Rme1 signal intensities normalized to intensities of HSP70. SGC3027: n = 11, four separate experiments, IC50 = 2.4 ± 0.1 µM; SGC3027N: n = 4 technical replicates, IC50 > 40 µM (mean ± SEM). Modified from PIMID:32409666. (Magdalena Szewczyk)
For experimental details please go to Zenodo.
