XIAP and its interaction with type I and type II receptors.
Having spent time over the past few weeks purifying all the components needed for a pull down with XIAP, I did the experiment that looked for interactions between XIAP and various type I and type II receptors in both the un-phosphorylated and phosphorylated form.
I used phosphorylated RIPK2 as a positive control as previous experiments in the lab have shown that XIAP binds to phosphorylated RIPK2.
XIAP was GST tagged so I used GST resin to bind to it, and then use various proteins as bait passed over the column to assess whether the two interacted.
I wanted to see whether phosphorylation influenced binding at all and so I divided my initial bait samples in half and used half untreated, and used the other half with 1mM ATP and 1mM Mg2+ for 30 minutes.
All fractions were collected and run on SDS-PAGE gels and run at 170V for 50 minutes.
After staining the gels this is what I found:
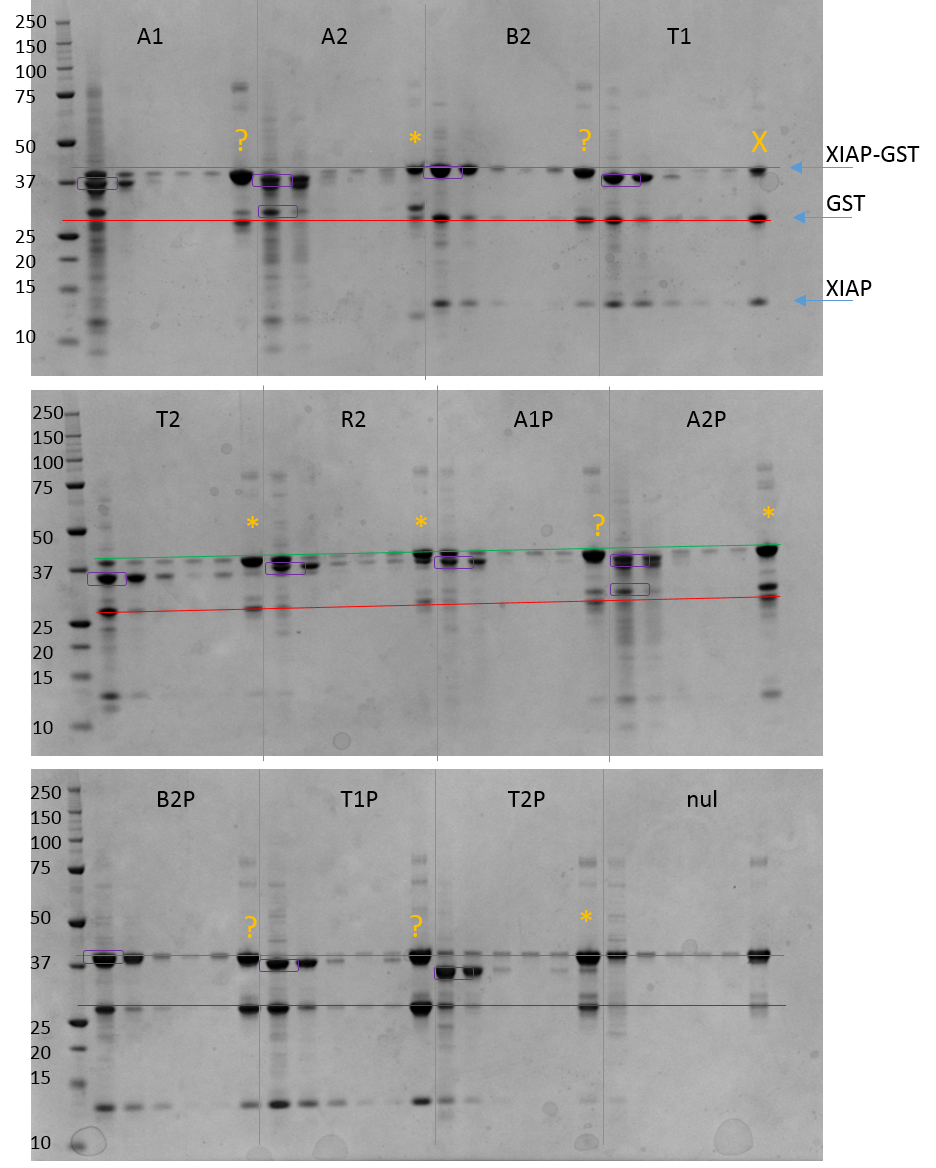
SDS-PAGE gel results for pulldown. Each protein has lanes 1 – 6.
1 = flow through of bait protein. 2 = wash 1. 3 = wash 2. 4 = wash 3. 5 = wash 4. 6 = Elution.
Some of the results are quite obvious and seem to show an association (ACVR2, TGFBR2), but others are less clear. My main problem is that XIAP-GST is around 41 KDa but Alk2 (39KDa), BMPR2 (40KDa) and Alk5 (39KDa) are all too close to XIAP-GST to distinguish on an SDS-PAGE gel. The wide bands in the elution fractions may suggest co-elution or it may just be large amounts of XIAP-GST.
The positive control (RIPK2) worked while the negative nul control (with no bait) also worked. I do note however that when XIAP is in the presence of other proteins there does seem to be some cleavage occurring as bands are seen at 28 and 14 kDa which correspond to the masses of GST and cleaved XIAP. This could be because the association with other proteins favours cleavage or it could be due to small amounts of contaminating Tev protease from the initial receptor purifications. Both these masses however are far enough away from the masses of the bait receptors to be easily distinguished.
To confirm the presence of the bait in the samples I can use mass spec to look for the receptor. I set up the samples ready to go but unfortunately overnight the mass spec machine suffered a pump error and my samples didn’t run. They’re still in the queue ready to run when the machine is fixed but I’m not sure how long that’s going to be. I shall report back when I know more!
Tm shift of M4K compounds with Alk2, Alk5 and RIPK2.
I also managed to run some Tm shift experiments with Alk2, Alk5 and RIPK2 against the M4K pharma. Some of these were repeats to ensure consistency across runs, while some of these were new.
The Tm shifts show that none of the compounds are particularly stand out for RIPK2 (only K4K2072 even gets close to being a significant Tm shift and even that is only 8°C). However a few compounds are seen as being fairly potent against Alk2 and Alk5 as shown by bars that go beyond the dotted line. Of these, there were a couple that seemed to show interesting selectivity for Alk2 vs. Alk5 (as shown in the scatter plot of Alk2 vs. Alk5 Tm shifts – those in the top right corner below the red line are of most interest.) – these show fairly good correlation to the in-vivo results Jong Fu is getting which is very reassuring.

Tm shift data of RIPK2, Alk2 and Alk5 against M4K Pharma compounds on plate K115
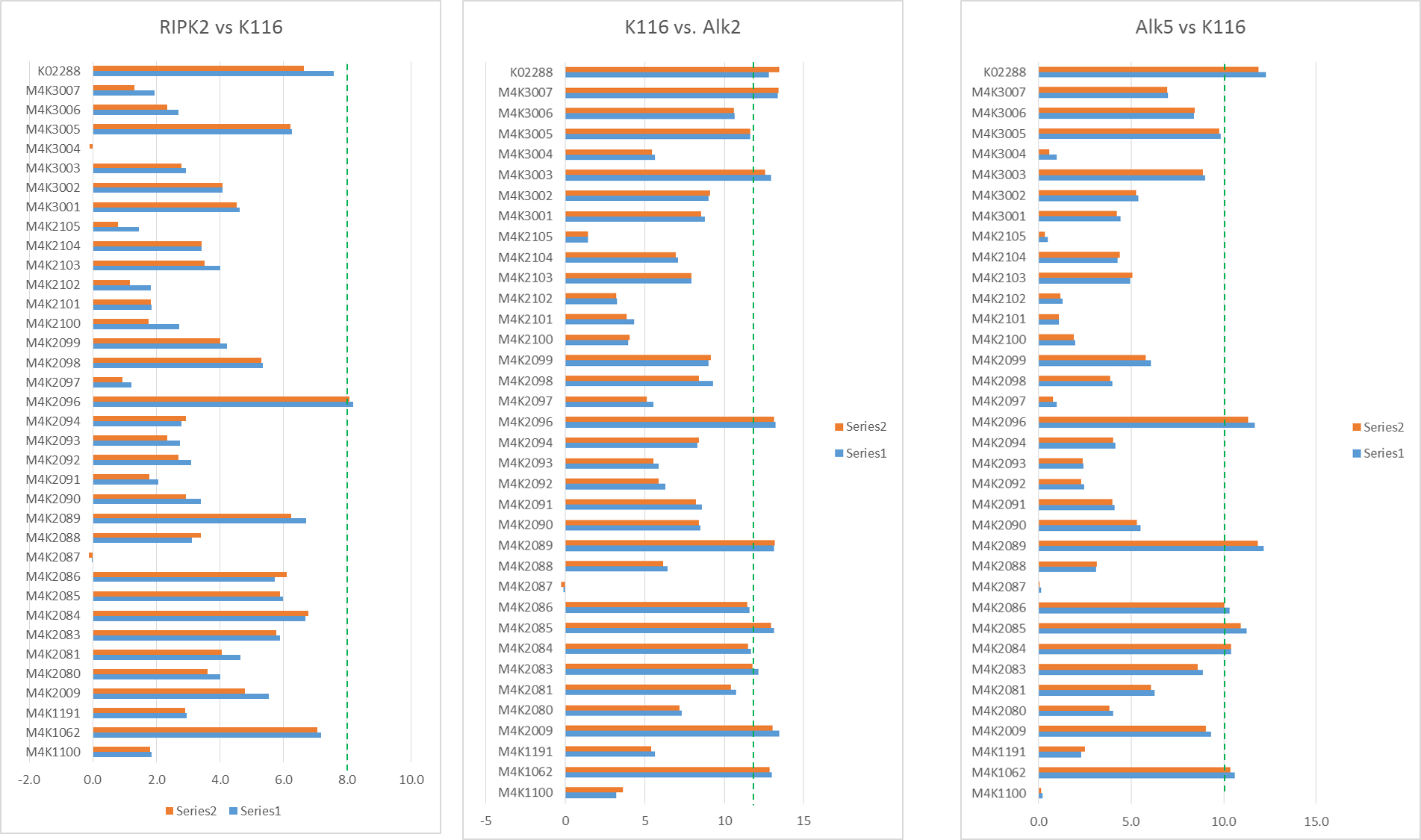
Tm shift data of RIPK2, Alk2 and Alk5 against M4K Pharma compounds on plate K115
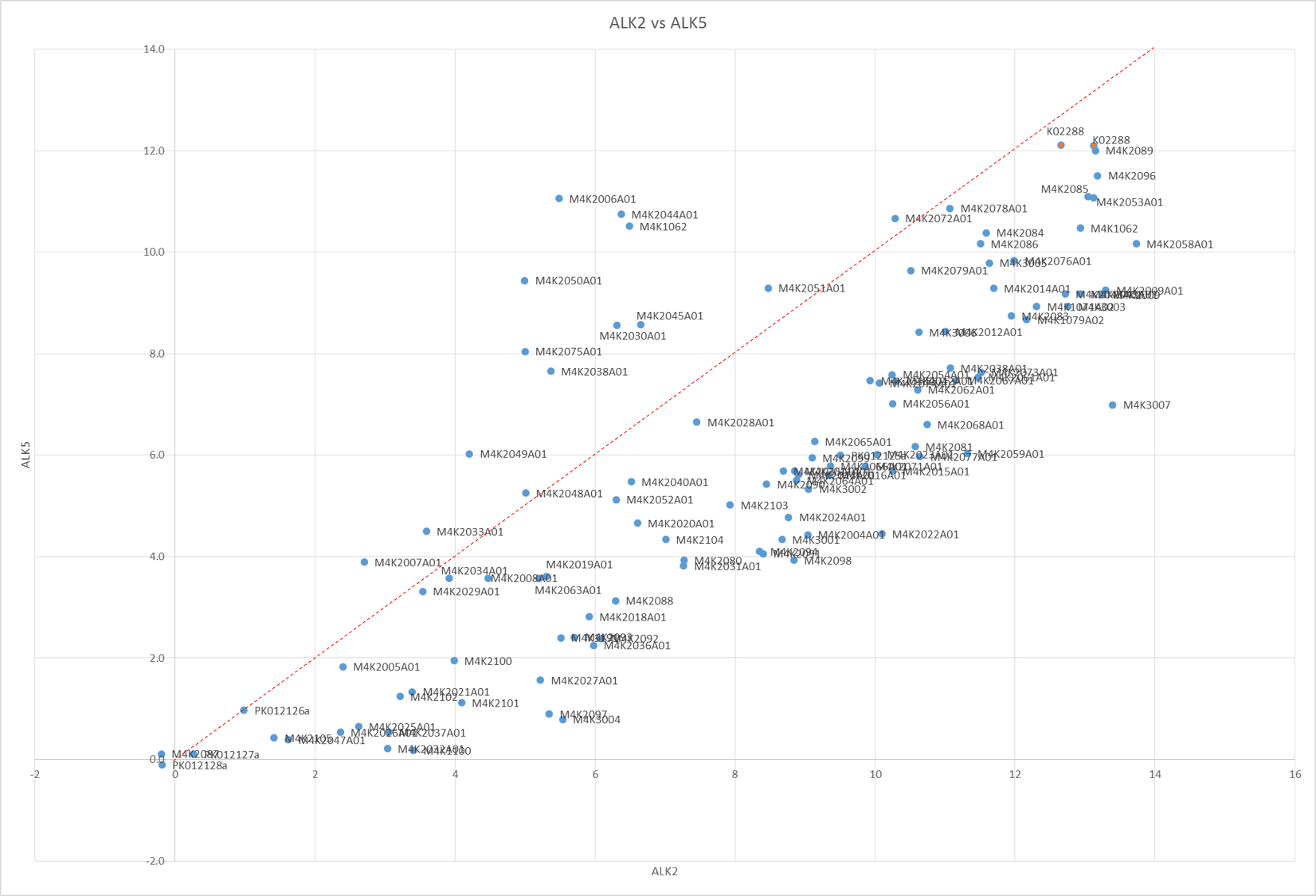
Tm shift data of Alk2 plotted against Alk5 of all M4K Pharma compounds on plates K115 and K116.
The next batch of compounds has just come in from M4K Pharma so this week I’ll be adding them to the Tm shift plates that I’ve got and running those experiments as well as following up on the pulldown results that I’ve got.
If you want to read more details of either of these experiments you can find details over on zenodo.
