SNS-032 is a CDK2 inhibitor with an IC50 of 48 nM in cell-free assay and is 10 and 20-fold selective over CDK1 and CDK4 respectively (http://www.selleckchem.com/products/SNS-032.html). In my previous post (https://openlabnotebooks.org/introduction-to-my-kcgs-cdk-family-project/) I mention the aim to develop a narrow profile inhibitors for understudied kinase. To do so I have synthesized SNS-032 analogues since SNS-032 inhibits a narrow family of interesting kinases and seems to have great structural properties suitable for cellular activity. These analogues have been prepared through a 2-step synthesis (Kimbal, S. D.; J. Med. Chem., 2004, 47, 7). First, reduction of thiazole 1 with sodium borohydride, followed by in situ alkylation with oxazole 2 provided amine 3 in excellent yield after purification (93%). Final compounds 4 were isolated by reaction of amine 3 with the corresponding carboxylic acid using HATU along with DIPEA, for the formation of the amide bond.
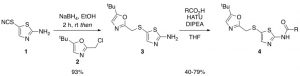
The compounds picked for the preparation of SNS-032 analogues are shown below. Note, compounds 12–14 were synthesized directly from SNS-032 in one step. To explore the amide group I introduced building blocks with different size, shape, and functionality. Our strategy is to make first relatively big changes on the molecule so we can have more of a chance to see modulation of the different kinase activities in the screening. Perhaps this is risky, but we feel it improves our chance of moving into new space selective for kinases of interest. Removal of the basic group of SNS-032 was accomplished with compounds 5–8, whereas compounds 9–11 introduced aromatic groups to look for different interaction with the enzyme. Compounds 12-14 kept the framework of SNS-032 but modified the basicity of the piperidine nitrogen and introduced some new functionality.
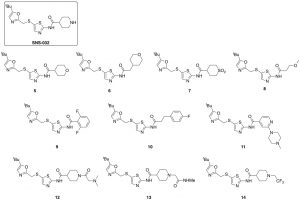
With the analogues in hand, compounds 5–14 were screened against a set of kinases targeted by SNS-032 (including some understudied kinases we are particularly interested in), and data is provided in the table below. There are many interesting results inhere for us to follow up on. From the panel screened, a small change from SNS-032 to compounds 5–7 provided a more selective profile. Also, removal of the cycle and introduction of a simple acyclic alkyl chain instead (compound 8), provide a CDK2 potency of 28 nM and the selectivity for CDK2 is improved, just like compound 14 provide a CDK2 selective potency of 58 nM. Aromatic functionality in compound 9 provided activity almost all across the panel, but the activity was totally killed with a more flexible compound 10. From the results with compounds 12, 13 and SNS-032, looks like it may be difficult to discriminate between CDK16, CDK17 and CDK18. We can look at the active sites to see what differences they have. One of the most promising results to me is with compound 11. Introduction of a more polar and soluble pyridine-N-methylpiperazine provided a narrow profile inhibitor getting activity on CDKL2 and CDK7, 63 and 43 nM, respectively. Many of the SNS-032 targets fell off significantly with this change. For example, CDKL5 went from 1.5 nM on SNS-032 to 2800 nM for compound 11. CDC2L2 went from 48 nM (SNS-032) to 8600 nM (11).
From these results, there are a few ways to follow up on this project. Although CDK2 is not as understudied as other kinases, compound 6 diminished activity across all the panel screened except with CDK2, giving a starting point we could use to further improve towards a CDK2 selective tool. Compound 11 showed a narrow profile with CDKL2 and CDK7 activity, and therefore I will move on to investigate the oxazole ring and thiazole core with the hope to find an improved and selective compound for either of these kinases. In the coming weeks I’ll show the chemistry to change the oxazole portion and screening data will come after that! Feel free to suggest further ideas and if you have any questions or comments or want more detail on anything, please let me know.

One Reply to “SNS-032 analogues update”