BRSK2 Compounds
NRF2 is a transcription factor that regulates the expression of antioxidant proteins and functions to protect against oxidative stress caused by inflammation. Under normal physiological conditions, NRF2 is degraded by KEAP1 and Cullin 3 (CUL3) through a process called ubiquitination (ubiquitin-dependent proteolysis). Following ubiquitination, it is transported to the proteasome where it is degraded, Figure 1.
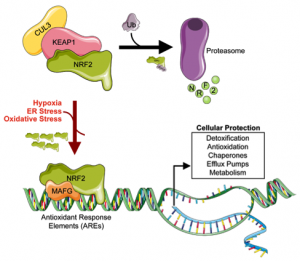
Figure 1: Ubiquitin-dependent proteolysis of NRF2
Under stress conditions (Hypoxia, ER Stress, Oxidative Stress), the ubiquitination process is disrupted leading to a build-up of NRF2, which translocates into the nucleus, forms a dimer with MAF proteins and binds to the antioxidant response elements (AREs) and initiates their transcription.
Balanced NRF2 signalling is required to maintain cell and tissue homeostasis, however alterations in NRF2 or KEAP1 or CUL3 is associated with diseases including premature aging, neurodegenerative disorders, chemical induced carcinogenesis, tumor progression, metabolic disorders and resistance to chemotherapy. Figure 2.
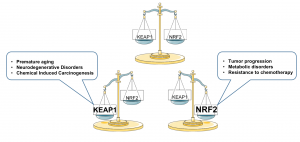
Figure 2: Balanced NRF2 signaling required to maintain homeostasis
BRSK2, a member of AMP-activated protein kinase (AMPK) subfamily of serine/threonine kinases is among kinases that repress NRF2 mediated transcription.
On this project, we are working with Dr. Ben Major (Dep’t of Cell Biology and Physiology, UNC) whose lab is actively engaged in the study of how perturbation of specific signal transduction pathways (specifically KEAP1/NRF2 signaling pathways) contributes to the initiation, progression and dissemination of diseases such as cancer.
Synthesis: Our initial synthetic efforts are focused on a number of carefully selected analogs that have been shown to be active BRSK2 inhibitors. The general synthetic route is provided below in Scheme 1, J. Med. Chem. 2013, 56, 1996-2015. The commercially available 5-bromo-2-fluorobenzaldehyde, 1, was converted to the corresponding 6-bromoquinazolin-2-amine, 2, following treatment with guanidine carbonate in DMA. The compound thus obtained, was able to undergo Suzuki coupling with 3-pyridinyl- or 4-pyridinylboronic acid to afford the amino derivative 3. Palladium catalyzed cross-coupling with a variety of arylbromides furnished the final aminoquinazoline analogs (compound 4, in Scheme 1).
Scheme 1: Synthesis of aminoquinazoline analogs
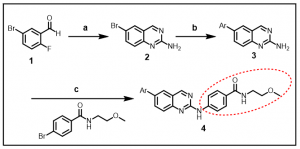
Ar = 3-pyridinyl or 4- pyridinyl; Indicated red circle: Introduction of diversity
Reagents and conditions: guanidine carbonate, DMA, 140 oC, 44%; (b) Pd(PPh3)4, K2CO3, ArB(OH)2, DME/water, 90 oC, 66%; (c) Pd2(dba)3, BINAP, CS2CO3, dioxane, 100 oC 54%.
A number of compounds have been submitted for biological evaluation and we are currently waiting on data that would inform our SAR studies on the design and synthesis of active BRSK2 inhibitor compounds.
