In a previous post, I outlined initial experiments to develop a differential scanning fluorimetry (DSF) assay for USP5 Zf-UBD where I measure the resistance of USP5 to thermal denaturation as small molecules bind and stabilize the protein. I previously could not observe any significant change in the melting temperature of USP5, even using a ubiquitin peptide that I know binds (though weakly) to USP5 Zf-UBD. As a follow up experiment, I tested if the presence of detergent in the buffer and if the addition of higher concentrations of ubiquitin RLRGG peptide resulted in a thermal shift of USP5 Zf-UBD. Experimental details are on Zenodo.
I was hoping that the presence of detergent in the buffer would prevent possible non-specific interactions and aggregation of USP5 Zf-UBD without significantly changing the signal by interfering with binding of the fluorescent dye to the unfolded protein. 0.01% Triton X-100, a non-ionic detergent does not significantly change the melting temperature of USP5 Zf-UBD in comparison to buffer conditions with no detergent but does increase the dynamic range of fluorescence measurements! Going forward, I will be including 0.01% Triton X-100 in my buffers for DSF experiments.
Unfortunately, there was no apparent stabilization of USP5 Zf-UBD in the presence of higher concentrations of a ubiquitin RLRGG-peptide, probably due to low affinity of the peptide to the protein domain. Next, I’ll be testing ubiquitin peptides of varying length against the USP5 Zf-UBD. Hopefully, one of these peptides will have a higher affinity for the protein and show a thermal shift with increasing peptide concentrations. This is the positive control I need before I can start using this assay to ask whether compounds bind USP5 Zf-UBD.
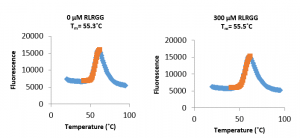
Figure 1. Representative regression charts of 30 µM USP5171-290 and [RLRGG peptide] in 100 mM Hepes pH 7.4, 150 mM NaCl, 1 mM TCEP, 0.01% (v/v) Triton X-100. There is no thermal shift of USP5 Zf-UBD with increasing RLRGG concentrations.
