In previous experiments, a FITC-RLRGG ubiquitin peptide had a Kd of ~55 µM for USP5 Zf-UBD. Different lengths of ubiquitin peptides were designed to see if increasing the peptide length resulted in a gain in affinity for the protein domain in the FP assay: LRLRGG, RAHGLRLRGG, RAHGRAKHGLRLRGG. The 6-mer peptide, LRLRGG, is the native C-terminal sequence of ubiquitin; it was hypothesized that the addition of a leucine residue would increase the affinity of the peptide to USP5 Zf-UBD (Figure 1).
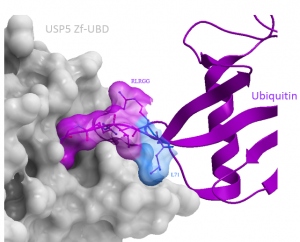
Figure 1. Interaction of ubiquitin LRLRGG with USP5 Zf-UBD (PDB: 2G45)
The 10-mer and 15-mer peptides were designed with small basic amino acids at the N-terminus to potentially improve binding properties of the peptide to the surrounding acidic surface of USP5 Zf-UBD as seen in red in Figure 2.
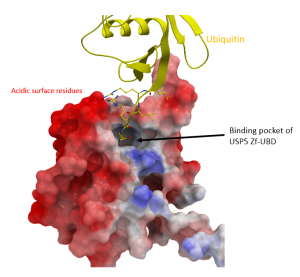
Figure 2. Surface model of USP5 Zf-UBD (PDB: 2G45) and ubiquitin
Experimental details can be found on Zenodo. Increasing the length of the ubiquitin peptide did not improve the affinity for the 6-mer, 10-mer or 15-mer peptide against USP5 Zf-UBD. This is likely due to lack of stabilizing interactions with other surface regions of ubiquitin. Unfortunately, it seems as though the FP competition assay with the ubiquitin peptides and the Zf-UBD of USP5 will be a no-go for screening libraries due to the weak affinity of the peptides against the protein domain and low FP dynamic range. I may have to use different protein constructs, such as the full length protein. I will be focusing on SPR and NMR assays to screen compounds against USP5 Zf-UBD; however, once I’ve established a screening assay, I will need a competition assay to confirm that compounds are displacing ubiquitin. If you have comments or suggestions please feel free to comment below or send me an email!

2 Replies to “USP5 Zf-UBD Fluorescence Polarization Displacement Assay #3”