For those following the USP5 project on my open notebook, you will recognize that assay development has been quite a challenge for screening ligands against USP5 zinc finger ubiquitin binding domain (Zf-UBD). In the past, I was unsuccessful in developing a differential scanning fluorimetry, and fluorescence polarization screening assay. I then tried 19F NMR spectroscopy screening, where ligand binding could be detected by structural perturbations in the resonance of a fluorinated tryptophan at the binding pocket. This assay works well, but so far, it has been more of qualitative than quantitative assay. I also used a surface plasmon resonance assay (SPR) to quantify and validate the binding of small molecule compounds to the protein domain of interest. Although I am able to measure binding constants with the SPR assay, it would be good to have a secondary assay that validates binding measurements. Also, SPR chips can be quite expensive so having a cheaper, higher-throughput assay would also be beneficial.
The medium-throughput 19F-NMR assay gave rise to the idea of exploiting the intrinsic fluorescence of aromatic amino acids of USP5 Zf-UBD for a quantitative screening assay. The three amino acids that are responsible for the intrinsic fluorescence of proteins are tryptophan, tyrosine and phenylalanine (Figure 1).
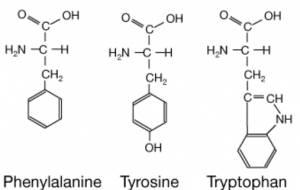
Figure 1. Chemical structure of aromatic amino acids
Aromatic amino acids have characteristic absorption and emission wavelengths, of which tryptophan has higher fluorescence than tyrosine or phenylalanine residues in a protein. There are 2 tyrosines and one tryptophan in the USP5 Zf-UBD pocket (Figure 2): theoretically, if a ligand is binding, we should see a change in the measured UV fluorescence intensity. Using USP5171-290, I attempted to develop a tryptophan fluorescence assay to detect ligand binding. You can find experimental details on Zenodo.
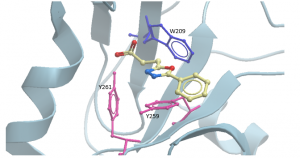
Figure 2. Aromatic amino acids in binding pocket of USP5171-290 with compound DAT180 (PDB: 6DXT)
As expected, as the concentration of USP5 Zf-UBD increases, the fluorescence intensity increases linearly (Figure 3). In other words, as the number of aromatic amino acids increases, the measured fluorescence intensity also increases.
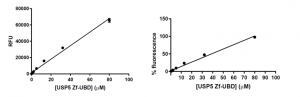
Figure 3. USP5 Zf-UBD fluorescence intensity measurements
Next, I tested compounds I had previously identified and co-crystallized in the USP5 Zf-UBD pocket, and others that bind to USP5 Zf-UBD according to the 19F NMR assay. Compound titrations were done in the presence and absence of USP5 Zf-UBD. Compound titrations in buffer only were used for blank subtraction to eliminate the fluorescence of the compounds from measured fluorescence of the protein + compound. Compound titrations with N-acetyl L-tryptophanamide (NATA) (Sigma Aldrich), a tryptophan analog was used as a control for the inner filter effect (IFE) (Figure 5).
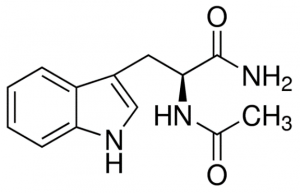
Figure 4. Structure of N-acetyl L-tryptophanamide (NATA)
The IFE is when the compound and protein both absorb at the excitation wavelength and the absorbance of the compound decreases emission of the protein. Theoretically, no change in fluorescence should be observed with compound titrations with NATA as the compounds are not expected to bind NATA. If a change in fluorescence is observed, this can be attributed to the IFE and corrected for using a correction factor.
The ubiquitin peptide, LRLRGG were used as positive control for USP5 Zf-UBD. Although there is a slight increase in fluorescence as the concentration of the ubiquitin peptide increases, binding affinity could not be measured (Figure 4a). This is discouraging because if I can’t measure the binding affinity of my positive control, it will be difficult to assess the accuracy of measured binding affinities of other ligands. Interestingly, while the ubiquitin peptide showed an increase in fluorescence, all the compounds showed a decrease in fluorescence as the concentration of the compounds increased (Figure 4b). I am not sure why there is difference in the change in the fluorescence of the peptides versus the compounds, as both the peptide and the compounds showed perturbations of the tryptophan in the binding pocket with the 19F NMR assay. Perhaps, the way in which the peptide binds to the binding pocket changes the environment of the hydrophobic amino acids in a different manner than when the compounds bind to the Zf-UBD.
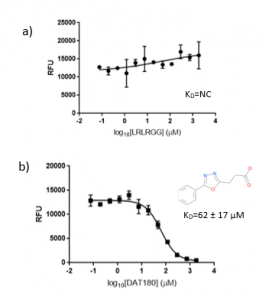
Figure 5. 1:2.5 12 pt dilution series of a) LRLRGG and b) DAT180 with 10 µM USP5171-290
The negative controls DAT199 showed no binding to USP5 Zf-UBD (Figure 6).
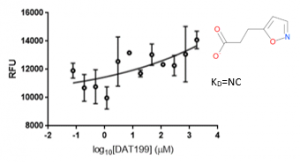
Figure 6. 1:2.5 12 pt dilution series of DAT199 with 10 µM USP5171-290
It looks like the tryptophan fluorescence assay can be used to detect ligand binding; however, it is difficult to conclude that the binding affinities of the ligands are accurate due the inability to measure the ubiquitin peptide as a positive control and the number of questions that have arisen. Anyone with expertise on this assay, could you please advise?
1) Why is there a difference in the change of fluorescence with ubiquitin peptides vs. small molecule compounds?
2) Was the IFE correction method used the right mathematical model for this assay?
3) Are the binding affinities measured for the small molecule compounds accurate, especially for those compounds that display the IFE?
As of right now, it doesn’t seem like I’ll be using this assay to screen compounds as I am uncertain how to interpret the data. In the future, I will be using a surface plasmon resonance (SPR) to measure the binding affinities of all these compounds. It will be interesting to see if the binding affinities measured by tryptophan fluorescence can be correlated to SPR binding affinities; if so, perhaps the tryptophan fluorescence assay will be a viable screening assay. Nonetheless, for USP5 I have already developed a 19F NMR spectroscopy assay for qualitative screening and SPR for quantitative validation, which seems to be working well.
For those familiar with designing fluorescence based assays and accounting for IFE’s please feel free to contact me if you have suggestions/advice on my development of a tryptophan fluorescence assay for USP5 Zf-UBD.
References
Epps, D., Raub, T., Caiolfa, V., Chiari, A., & Zamai, M. Determination of the Affinity of Drugs toward Serum Albumin by Measurement of the Quenching of the Intrinsic Tryptophan Fluorescence of the Protein. J. Pharm. Pharmacol, 1999. 51: 41-48.
Fonin, A., Sulatskaya, A., Kuznetsova, I., Turoveroc, K. Fluorescence of Dyes in Solution with High Absorbance Inner Filter Effect Correction. PLoS ONE, 2014: 9(7).
Wang, T., Zeng, L., & Li, D. A review on the methods for correcting the fluorescence inner-filter effect of fluorescence spectrum. Applied Spectroscopy Reviews, 2017: 52 (10).
