In my previous post, I confirmed that the two active site fragments (fragments 1 and 2) and the gating loop fragment (fragment 5) bind to and inhibit HAO1 in solution. Next, I chose some larger compounds to test by entering each fragment hit structure into a search engine (emolecules.com) to look for commercially available compounds that include the fragment within their structure. This resulted in 40 compounds based on the active site hits and 110 compounds expanding the gating loop hit.
Once the compounds arrived, I characterised them using the fluorescence-based (Amplex Red) activity assay that I described in my last post. After narrowing down my number of ‘good’ compounds by performing the assay at a single concentration (1 mM), I measured inhibition of these good compounds at a range of concentrations to determine their IC50 values – the concentration (of compound) at which the protein’s activity is reduced by 50% – to find out which compounds were most potent. I was able to determine reliable IC50s for 5 active site and 8 gating loop compounds (figure 1). You can find the experimental details in my Zenodo post here.
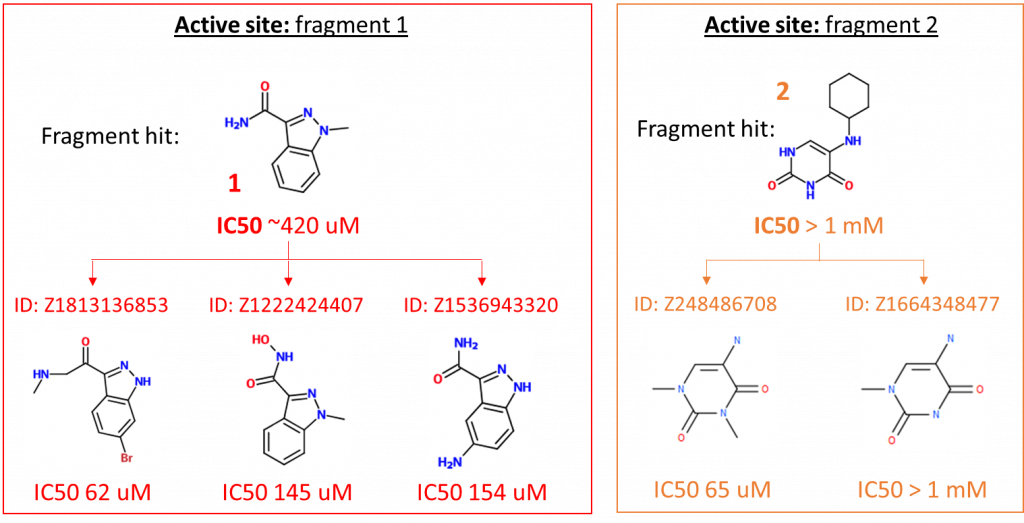
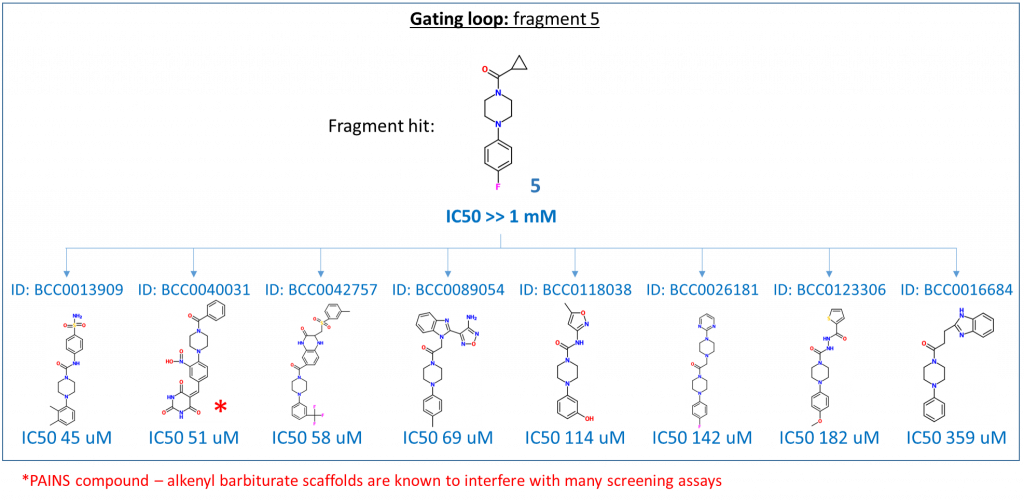
Figure 1: Structures and IC50 values for most potent follow up compounds expanding fragments 1, 2 and 5.
The three ‘take-home’ messages from this assay are:
- Potency of fragment 1 can be increased 10-fold by growing the fragment in several directions – this indicates considerable potential for increased potency by growing in multiple directions at once.
- Potency of fragment 2 can be increased >16-fold by removing the cyclohexane portion of the fragment. Such a high IC50 for so small a compound is very surprising – and very exciting!
- A massive increase in potency (greater than 20-fold) can be achieved for fragment 5 by growing in either/both directions. It is encouraging that the most potent compounds are those that expand the core hit from both ends of the structure i.e. In towards the active site and out towards the protein surface.
The next steps for this project are:
- Chemistry to make some follow-up (larger) compounds based on fragment 6, which binds at the HAO1 oligomer interface.
- Test how the follow-up compounds for the oligomer interface behave in solution.
- Use surface plasmon resonance (SPR) to measure how tightly follow up compounds bind to HAO1. Particularly important for the active site fragment 2 compounds that are surprisingly potent for such small fragments (to ensure they are ‘real’ inhibitors and not e.g. aggregating the protein)
- Soak the most promising compounds into HAO1 crystals to look at how they bind and help rationally design the next round of compounds.
