One of the questions to be asked when looking at the mutation in ALK2 that causes FOP, is how exactly does that mutation cause the disease. ALK2 we know is part of the BMP signalling pathway responsible for bone formation and the mutation results in excess bone formation – thus we can summarise that somehow the mutation leads to increased signal. The question is how?
Alk2 is known to phosphorylate SMADs 1, 5 and 8 (summarised as SMAD1/5/8) in order to signal downstream in the BMP signalling pathway. The instigation of this signal comes from the binding of an external BMP ligand to the domains of Alk2 and a type II receptor on the outside of the cell. This co-locates the internal domains (which incidentally is also where the Alk2 mutation is located) and seems to drive signalling.
I have been looking at the activation of SMAD1 by ALK2 in the presence or absence of a Type II receptor and in the presence or absence of the FOP ALK2 mutation. The activation of SMAD1 results in its phosphorylation which can be monitored by mass spectrometry.
- We know from previous experiments that Alk2 alone cannot activate SMAD1 even if the mutation is present, thus the Type II receptor is needed.
- We know that Type II activity isn’t necessary for SMAD1 activation as using a kinase dead version of BMPR2 still works.
- We know from previous experiments that you need both the kinase and GS domains of Alk2 in order to get SMAD1 activation.
- We know from previous experiments that BMPR2 and ACVR2 both work to allow SMAD1 activation.
- We know that Alk2 autophosphorylates its self to some degree, focused on the GS loop.
- We know that the type II receptor transphosphorylates Alk2 adding additional phosphorylation sites.
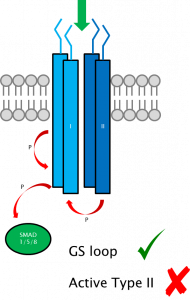
Interaction of the type I and type II receptors in the membrane.
So what don’t we know?
- We don’t know how the mutation in Alk2 effects the rate of SMAD1 phosphorylation
- We don’t know how transphosphorylation effects the rate of SMAD1 phosphorylation.
- We don’t’ know how the mutation effects the rate of autophosphorylation of Alk2.
This experiment can be performed using isolated purified proteins. I’ve been busy purifying Alk2 (wild type and two mutant versions – Q207E and R206H), ACVR2 and BMPR2. I’m now ready to combine these with samples of the Kinase Dead version of BMPR2 and SMAD1 that I have in the freezer into a mass spec based assay.
I mixed all the components together at appropriate concentrations, in this case all proteins were at 40uM, ATP was at 0.5mM, Magnesium ions were at 5mM, Manganese ions were at 2mM and DTT (a reducing agent to stop the proteins precipitating) was at 5mM. These concentrations are needed because if you drop lower, you lose the association between the type I and II receptors. In the cell they’d be brought together in the membrane by a ligand binding outside the cell which would co-locate the inner domains. Since we’re only looking at the inner domains in vitro, we need to artificially increase the concentrations to allow them to interact. 40uM was previously determined as being the lowest concentration at which this interaction can reliably be measured.
Once the ATP had been added, this started the phosphorylation reaction. I took a small sample at various time points (30s, 1min, 90s, 2min, 150s, 3min, 4min, 5min, 6min, 7min, 8min, 9min, 10min, 15min, 20min 25min) from my main reaction mix, and acid quenched the reaction in this sample to stop it going any further. This was done by diluting the sample in mass spectrometry running buffer containing 0.1% formic acid.
The samples were run overnight on the mass spectrometry machine to measure the mass of each of the proteins and the masses of any phosphorylated proteins. The peak heights for each of the phosphorylated states (I looked at up to 2 phosphorylation states for SMAD1 and 8 for Alk2) was extracted from the data and used to monitor the rate of phosphorylation of both Alk2 and SMAD1 over time.
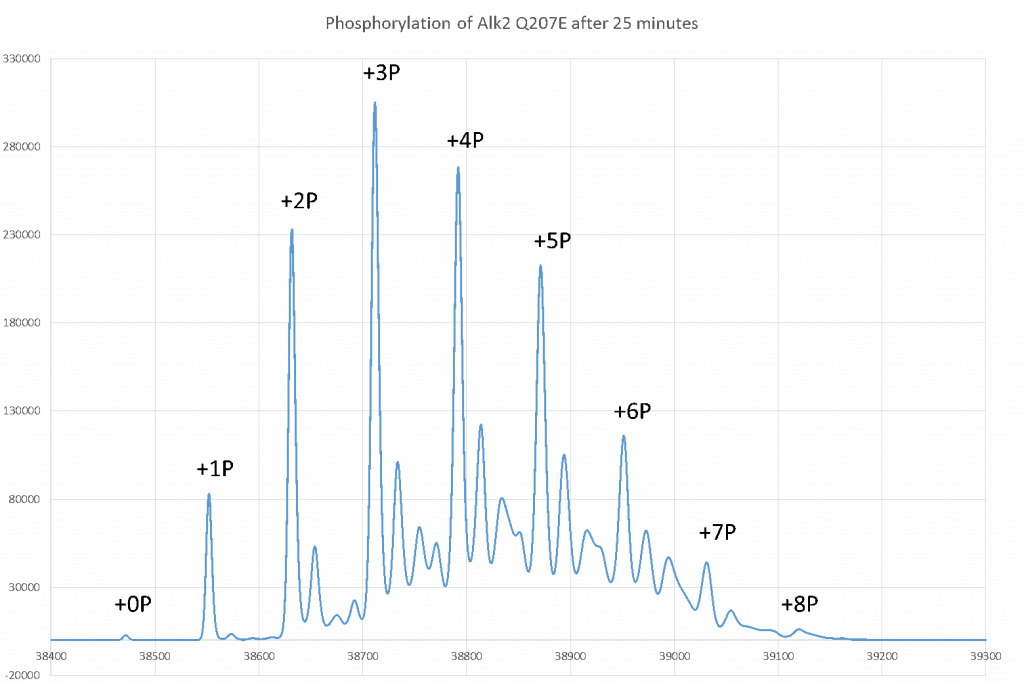
Total phosphorylation of Q207E after 25 minutes incubation with ACVR2.
I used the following combination of proteins in my reaction mixes:
| ACVR2 active | BMPR2 active | BMPR2 kinase dead | |
| Alk2 Wild type | X | X | X |
| Alk2 R206H mutant | X | X | X |
| Alk2 Q207E mutant | X | X | X |
This gave me the following results: In each case the important bit to look at is the initial slope of the reaction, not the absolute magnitude as that is more subject to Mass Spec variance.
Phosphorylation on SMAD:
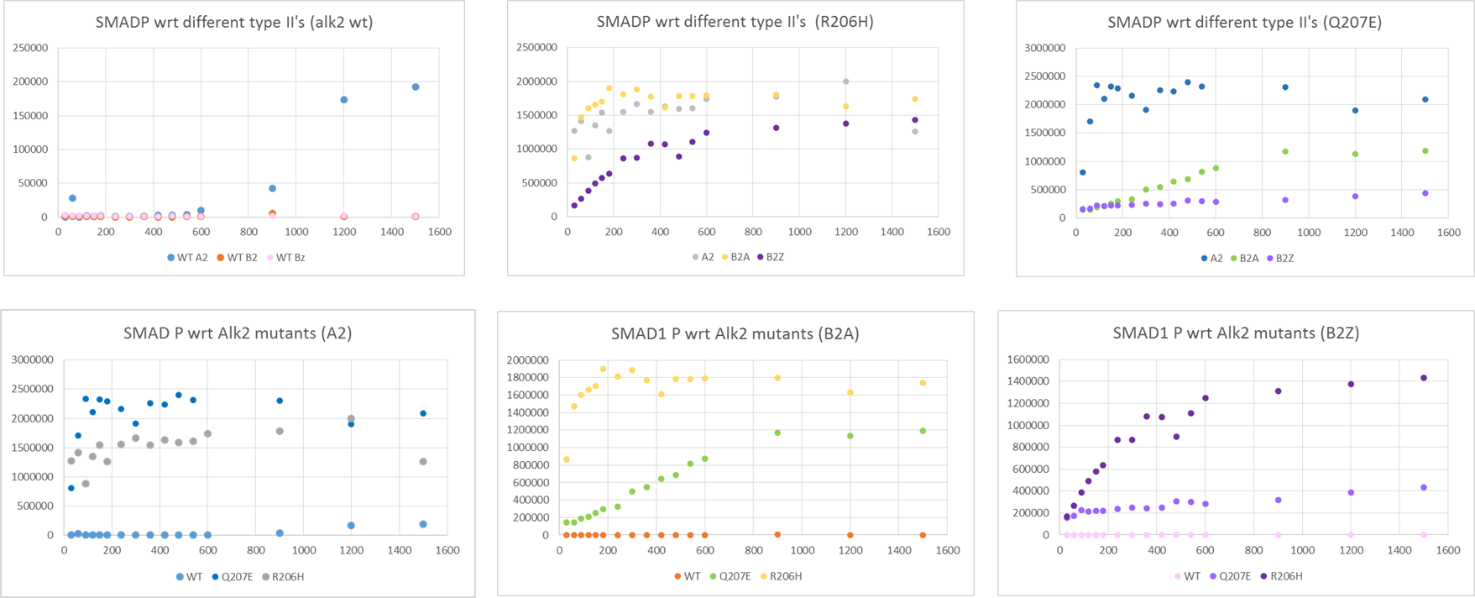
Graphs of SMAD1 phosphorylation comparing the effect of different Alk2 forms (WT, Q207E and R206H mutations) and different type II receptors (ACVR2, BMPR2 active and BMPR2 kinase dead)
Phosphorylation on Alk2
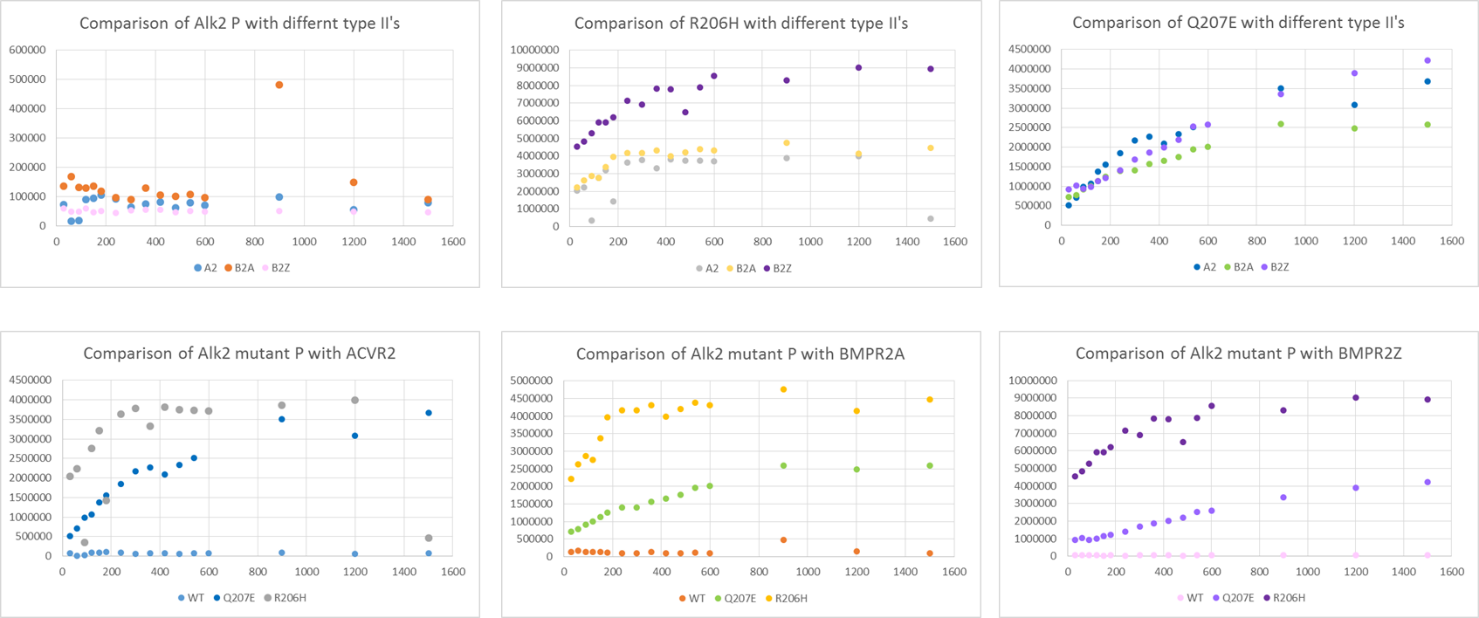
Graphs of Alk2 phosphorylation comparing the effect of different Alk2 forms (WT, Q207E and R206H mutations) and different type II receptors (ACVR2, BMPR2 active and BMPR2 kinase dead)
Interpreting this data is quite tricky because you can look at phosphorylation on either protein while at the same time comparing the effects of the Alk2 mutations or you can compare the effects of the different type II receptors. I haven’t got enough data to be able to get any quantitative data on initial rates of reaction but the trends shown in this experiment match the trends seen in the previous experiment I ran of this type some time ago (before I started writing this blog).
So, what can we conclude from this experiment?
- SMAD1 phosphorylation is slower with wild type Alk2 than with either of the mutations (levels out at 20 minutes compared to 5 minutes aprox.)
- When looking at SMAD1 phosphorylation, BMPR2 is a less good companion than ACVR2 when wild type Alk2 is used. Q207E somewhat counteracts this effect and R206H has the most pronounced counteraction bringing the rate of phosphorylation with BMPR2 to nearly equivalent to ACVR2.
- In general the Q207E mutation is a greater facilitator of SMAD1 phosphorylation than wild type. The R206H mutation is better than both of them.
- The rate of ALK2 phosphorylation is not effected by the type II protein used.
- The rate of Alk2 phosphorylation does change depending on which construct is used. WT has the lowest rate, Q207E has a faster rate while R206H has the fastest rate.
- The kinase dead version of BMPR2 leads to minimal difference in Alk2 phosphorylation but does reduce SMAD1 phosphorylation when compared to the BMPR2.
Next step: Try and repeat it again, however for that I need more wild type protein and we still don’t have the expression system back up and running for that.
For more details please look at the Zenodo entry for this pos
