In order to develop ligands addressing different possible binding sites of the transcription factor brachyury, fragments co-crystallized with this protein should be developed into ligands by using the program SparkTM (Cresset®).
Brachyury (T, TBXT) is an essential regulator of the notochord development. Brachyury is primarily expressed in the embryo but not in the majority of normal adult tissue. It is known that brachyury is overexpressed in chordoma, a primary bone cancer, and plays a role in the disease pathogenesis. Chordoma affects the skull-base, the mobile spine and the sacrum. It is a slow-growing, malign cancer that is locally invasive and has a tendency to recur although a surgical and/or radiation therapy was conducted.1
It is hypothesized that chordoma is originated from embryonic notochordal remnants. Brachyury is proposed to act as an aberrantly activated transcription factor and thereby be oncogenic. No approved targeted therapies, chemotherapies or immunotherapies are available for chordoma and no small-molecule inhibitors for brachyury have been developed.1 Therefore, a fragment-based approach should be explored to develop ligands for brachyury to study this protein.
Our colleagues Joseph Newman and Opher Gileadi at SGC Oxford co-crystallized wild-type human brachyury protein with various fragments which bind to the sites A-E (Figure 1, left). Thankfully, they provided the fragment and protein structures to us so that we can explore the usage of the SparkTM growing approach to develop ligands for these binding sites.
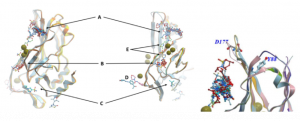 Figure 1. Left: Aligned X-ray structures of the human wild-type brachyury protein in complex with fragments at different binding sites (A-E). These first crystallization structures contain Cd2+ions (green balls). Right: Binding site A close to loop with D177 and in proximity to residue Y88.2
Figure 1. Left: Aligned X-ray structures of the human wild-type brachyury protein in complex with fragments at different binding sites (A-E). These first crystallization structures contain Cd2+ions (green balls). Right: Binding site A close to loop with D177 and in proximity to residue Y88.2
First, we focused on binding site A since most fragments bind to this binding site and site A is close to a loop that is mutated in chordoma-linked variant G177D3 and to the amino acid residue Y88 that, if mutated, affects binding to effector proteins4 (Figure 1, left).
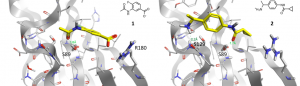
Figure 2. Fragment 1 and 2 co-crystallized in the binding site A of wild-type TBXT.
21 fragments were co-crystallized to binding site A. In Figure 2, the binding modes of two co-crystallized fragments are displayed. Fragments 1 and 2 show interactions or close proximity to two of the three amino acids Ser129, Ser89 and Arg180. Therefore, the growing approach of the program SparkTM should be used to identify new ligands that are able to form all three interactions. One growing approach started from the amino group of fragment 1 and was guided by fragment 2 towards the region of its carboxylate group (Figure 3).
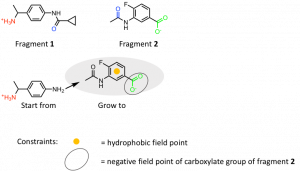
Figure 3: SparkTM growing approach starting from fragment 1 towards Arg180 guided by fragment 2. The functional groups of fragments 1 and 2 revealing H-bond interactions with amino acid residues are marked with the following colors: Ser129 (red), Ser89 (blue), Arg180 (green).
To achieve new ligands with similar properties in certain areas so-called field points were used. They are ‘condensed representations of electrostatic, hydrophobic and shape properties’ (Cresset®) of those areas where they are placed by the program SparkTM. Regarding the growing approach of fragment 1 and 2 the hydrophobic field point of the phenyl ring and the negative field point of the carboxylate group of fragment 2 were used. In total three of the 21 fragments bound to site A were used as starting points and each of them were grown in the area of two different fragments. The results were clustered by SparkTM depending on their structural similarity. Those compounds with the highest score of the first 20 clusters ranked according their scores were chosen to analyze their interactions by docking into binding site A using the docking program FlareTM (Cresset®). 4,000 docking results were inspected visually and compounds with a high docking score, high electrostatic complementarity and high number of interactions to the binding site were picked.
Figure 4 shows the docking pose of the example compound 3 that was obtained by docking 3 in binding site A of brachyury using FlareTM. Besides other criteria like docking score and electrostatic complementarity following interactions of 3 are promising for the co-crystallization of this compound with brachyury: two H-bond interactions of the positively charged amino moiety to Ser129 and His126, the proximity of the amide carbonyl-group to Ser89 and the H-bond interactions of the oxazole O-atom and N-atom towards Arg180.
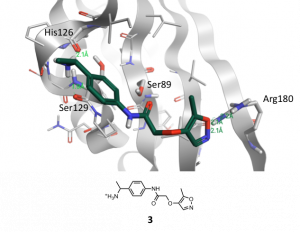 Figure 4. Binding mode of new compound 3 obtained by docking into binding site A of wild-type brachyury with FlareTM. H-bond interactions are marked with green dotted lines.
Figure 4. Binding mode of new compound 3 obtained by docking into binding site A of wild-type brachyury with FlareTM. H-bond interactions are marked with green dotted lines.
Additionally, new compounds were obtained by searching in the Enamine REAL database for similar compounds to those compounds that were found by the SparkTM approach and that showed promising docking results in FlareTM.
A selection of compounds that are identical or slightly different than those found in the SparkTM approach as well as compounds that represent similar ones to the SparkTM hits were purchased and will be tested regarding their ability to co-crystallize with brachyury. If the compounds bind to this transcription factor, they will represent promising starting points for future ligands of brachyury.
(1) Sharifnia, T., Med. (N. Y., NY, U. S.), 2019, 25, 292-300.
(2) Joseph Newman and Opher Gileadi, SGC Oxford, University of Oxford, UK.
(3) Pillay, N., Nat. Genet., 2012, 44, 1185-1187.
(4) Beisaw, A., EMBO reports, 2018, 19, 118-134.
