The molecular blueprint of the living organisms, deoxyribonucleic acid (DNA), is often wrapped with a family of proteins, called histones, in mammalian cells. The functions of histones are vast, including the protection of DNA, and most importantly, the regulation of gene expression. However, the details of gene regulation function of histones are not clearly understood, at least in terms of the helper molecules involved in this process.
We focus on a specific protein called SETDB1, which chemically modifies a certain part of the histones, causing the formation of a gene silencing signal (called H3K9me3). This gene silencing signal, which is a form of histone code, plays a pivotal role in the pathogenesis of various diseases, including many cancer types. We are motivated to find out a way to inhibit the abnormal functioning of SETDB1 that contributes to the progression of certain diseases. Our approach to do so is to solve the three-dimensional structure of SETDB1, which will help pharmaceutical researchers to design highly potent drug molecules.
As a general strategy, we first had a closer look at the domain organization of SETDB1, which also shows the functional layout of the protein (as shown in the figure below). SETDB1 is a large protein with a molecular weight of 143.1 kDa (approximately 400 times of table sugar, sucrose). From a structural as well as functional perspective, one side of the protein is studied very well. (See the blue boxes, called Tudor domains) However, the other sides are relatively unknown.
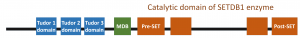
Figure 1. How SETDB1 looks like when we map out its functional units.
The aim of this project is to gather the structural information of relatively less resolved regions of the SETDB1 protein, which perform the catalytic activity. To achieve our goal, we will design new clones, expressing different regions of the protein and optimize their protein purifications. To keep you posted, we will share essential data and experimental design strategies that we develop here in this blog in the future. As always, we love to hear your thoughts.
