I wanted to determine whether USP5 Zf-UBD is a monomer or a dimer in solution as some known USP5 structures have shown that there is a cysteine surface residue that is not involved in Zn-coordination but forms disulfide bonds (2G43, 2G45, 3IHP). I used size exclusion chromatography with multiple angle light scattering (SEC-MALS) to determine the molar mass of the protein domain. MALS measures the light scattered by the protein at a range of angles allowing for extrapolation to determine molecular weight using a refractive index (RI) detector. For details of this experiment see the accompanying Zenodo record. The average calculated molar mass of the USP5 Zf-UBD was determined to be ~18 ± 2 kDa (Figure 1).
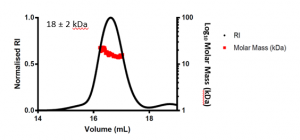
Figure 1. SEC-MALS of USP5 Zf-UBD171-290
The expected molar mass of the USP5 Zf-UBD monomer is ~13.5 kDa. The signal to noise ratio for the scattering was high due to the small size of the protein domain and the limited dynamic range of the MALS detector. Due to the high signal: noise ratio, the calculated molar mass is higher than expected at ~18 kDa; however, it is important to note that this is an average of the scattering across the USP5 Zf-UBD elution peak, where scattering is variable at the ends of the peak. It is the centroid molar mass of USP5 Zf-UBD that we are interested in. Visual inspection of the scattering at the centre of the USP5 Zf-UBD elution, where it is most consistent and has less signal: noise shows a molar mass of ~15 kDa (Figure 2). This is close to the expected molar mass of the USP5 Zf-UBD monomer.
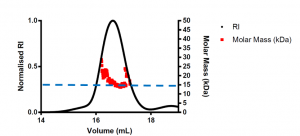
Figure 2. SEC-MALS of USP5 Zf-UBD171-290 scaled to see differences in scattering signal across the elution peak
Therefore, USP5 Zf-UBD is a monomer in solution. This is good news for when I begin to test compounds against this protein domain. Binding of ligands and assay signals would be affected if USP5 Zf-UBD was a dimer in solution.
