Hello everyone and happy New Year! I really, really wanted to end the year on a high note in December before I went off on holiday, but as it turned out, I didn’t have enough time to set up 18 plates and write a blog post on the last day. So I’ll just have to start the year on a high note.
I’m very happy to say I got another ACVR1/ALK2 structure in December, at 2 Å resolution, co-crystallised with M4K2158 (see images below, and the .mtz autoprocessed file is available here on Zenodo). The structure is not fully refined yet, but what’s interesting is the solvent side of the molecule is being stabilised by either a water contact (Chain A) or Asp293 (Chain B). This gives it really good density.
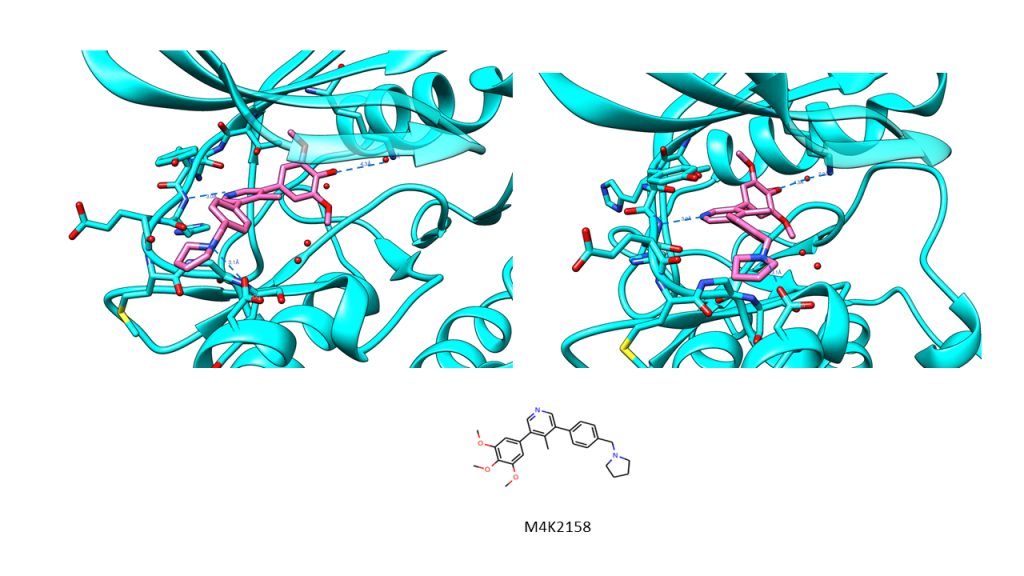
ACVR1 bound to M4K2158 at ~2 Å, showing interactions with His286 and a water at the back of the pocket
I also seem to have some new pretty hits with M4K2118, M4K2149 and M4K3003 (crystal pics below). What’s interesting about the M4K3003 hits is that I’ve really struggled to get crystals with this compound (as well as M4K3007 and 3010), and it is a pretty desirable structure. These hits came about as a result of a mistake, where I somehow managed to mix up my tubes and put ACVR1 with M4K3003 into my mega composite screen without seeds, and TNIK into a fine screen I had intended for ACVR1 with seeds. I only discovered this because I mass specced the TNIK just before setting up the plates, then rushed off to do them as it was the SGC Christmas party that night and I needed to finish (best not to rush experiments). When I checked the mass spec result later, I got the exact mass of ACVR1! So I went back to the crystallography room and recovered what protein I could from the source plate for the TNIK plates, and mass specced it again. Same result – ACVR1. But when I looked at the drop images for those plates (now containing ACVR1 + M4K3003), I found a bunch of crystals! Happy serendipity!! These will go on the trip to Diamond on the 21st of January and then we’ll know whether the mistake really ended well or not. Fingers crossed!
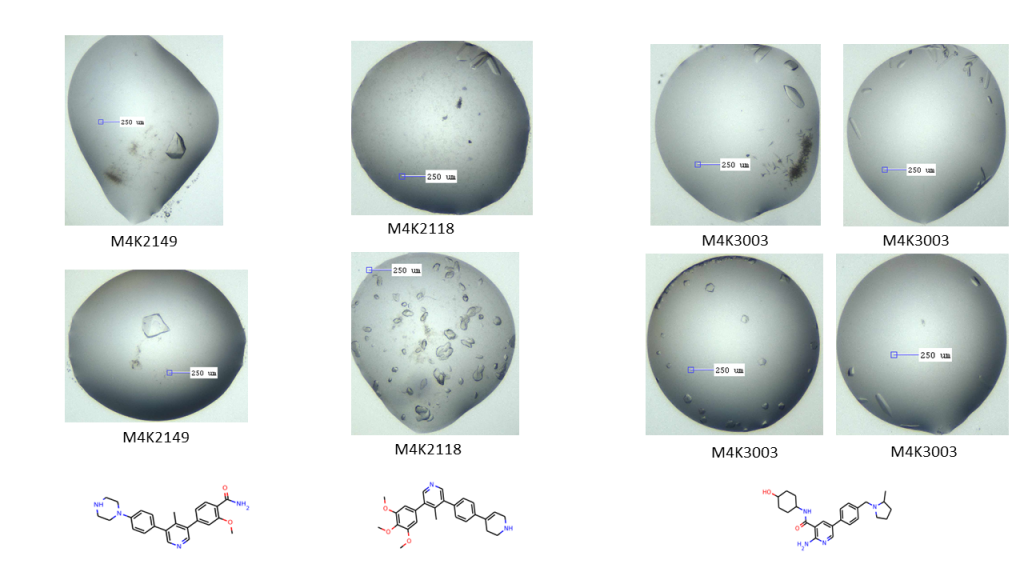
ACVR1 co-crystallised with M4K2149, 2118 and 3003 – hits to send to the synchrotron
You may well ask, why was I setting up plates with TNIK? As it happens, some of our collaborators’ assays show M4K2009, our main compound of interest, hits both TNIK and DDR1 with pretty high affinity. We’ve been struggling to get crystals of ACVR1 with M4K2009 – for whatever reason, it doesn’t want to make crystals. So it seemed like a good idea to see if TNIK or DDR1 will co-crystallise with it. TNIK (TRAF2 and NCK Interacting Kinase) is a serine/threonine kinase that functions as an activator of the Wnt signalling pathway. DDR1 (Discoidin Domain Receptor Tyrosine Kinase 1) is a tyrosine kinase that functions as cell surface receptor for fibrillar collagen and regulates cell attachment to the extracellular matrix, remodelling of the extracellular matrix, cell migration, differentiation, survival and cell proliferation (according to its GeneCard).
While I was about it, I thought it would be useful to set up plates with BMPR1A (ALK3), for the same reason I set up TGFBR1 (ALK5) before – to see what off-target structures look like with some of these compounds. The purification protocols for TNIK and BMPR1A were as standard, but they’re available here on Zenodo.
Some other good news is that I’ve figured out how to reproduce my XChem crystals fairly reliably. On the last day before the holidays I set up 6 plates of ACVR1 with M4K2117, which gives really beautiful, well-diffracting crystals. I used 10, 12 and 15 mg/ml protein, and made plates with and without seeds. Well, the ones without the seeds grew fabulously, within 2 weeks, and the ones with seeds didn’t! The next step with these then is to test what happens if I soak them in different formulations of lower salt with and without some PEG3350. That’ll be for next week. Have a great weekend!
