In my previous post, I introduced our fragment screening results on HAO1. Specifically, I told you that we identified five promising fragment hits, from a little over 400 collected structures, clustering to three biologically relevant sites: the active site (fragments 1 and 2), the gating loop (fragment 5) and the subunit interface (fragments 6 and 7). In this post, I will go into a bit more detail about what these fragment-bound structures inform us about HAO1 as a drug target. You can find out more in the Zenodo document here.
Previous HAO1 structural information is important for drug discovery
Two key features of the structure of HAO1 are important for the understanding of our data: the gating loop (figure 1A) and tryptophan 110, an active site residue that coordinates the substrate (figure 1B). Both features contribute to the biochemical characteristic of HAO1: it can metabolise a broad variety of substrates, ranging from the 2-carbon glycolate to the 16-carbon 2-hydroxypalmitate.
The gating loop (aa 169-212), often referred to as ‘loop 4’ due to its position between the fourth beta sheet and the fourth alpha helix, is highly flexible, as demonstrated by the variable orientation and degree of disorder observed in the published crystal structures, and is poorly conserved in sequence between members of the hydroxy-acid oxidase protein family.
Similarly, tryptophan 110, in contrast to the other residues in the active site, varies between family members: enzymes that prefer bigger substrates have a smaller residue at this position. In HAO1, tryptophan 110 is very flexible and can either point in towards the active site to metabolise the very small substrate glycolate or rotate outwards to bind larger substrates (or inhibitors). For a more detailed explanation of these structural features, including the movement of tryptophan 110 upon inhibitor binding, check out this paper.
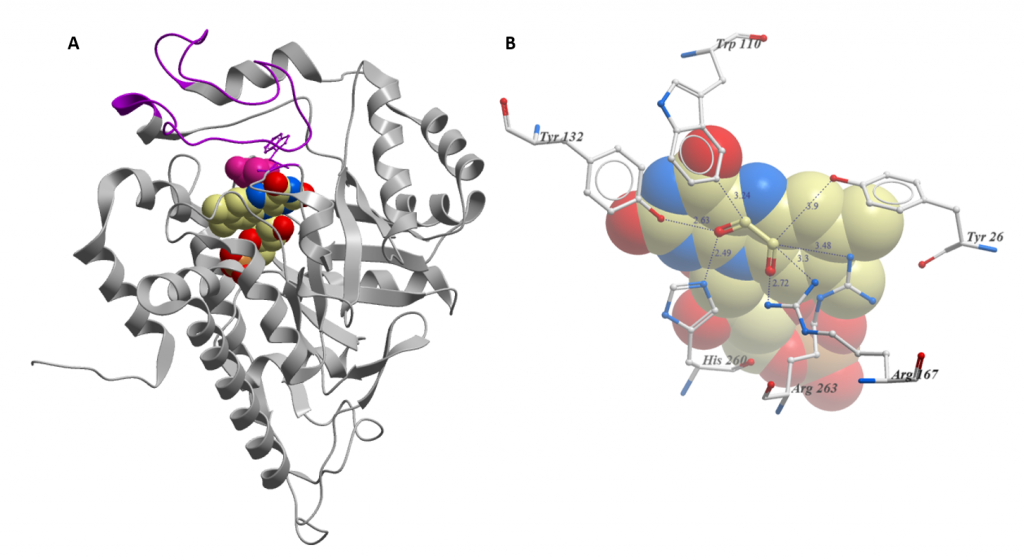
Figure 1: Key structural features of HAO1 (PDB code 6GMB). A. Overall structure: FMN is shown as spheres coloured by atom type; Glycolate is shown as pink spheres; and gating loop is coloured purple. B. Close-up of active site with FMN and glycolate bound. Residues that coordinate the substrate are shown as sticks. Distances from glycolate are in Angstrom. FMN is shown as spheres.
HAO1 shows considerable binding plasticity
The active site (fragments 1 and 2) and gating loop (fragment 5) fragments have an induced fit binding mode, causing conformational changes in nearby residues and, most notably, in the gating loop when they bind (figure 2). We can exploit this binding site plasticity when designing optimised inhibitors.
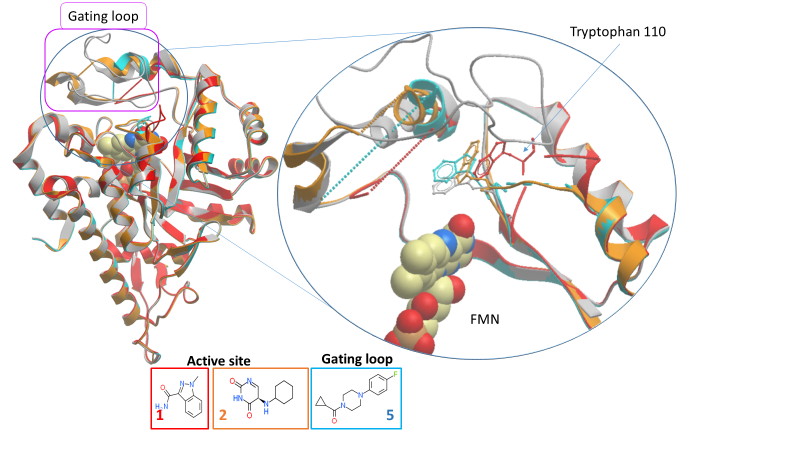
Figure 2: Movement of gating loop and key active site residues by fragments 1, 2 and 5 (PDB IDs 5QIH, 5QIB and 5QIC respectively).
Analysis of the active site hits (fragments 1 and 2)
Fragment 1 contains a polar head group attached to a five-membered ring, similar to published inhibitors. However, the binding position of fragment 1’s five-membered ring is different and so may still yield novel inhibitors.
The structure with fragment 2 is a little puzzling as only the unsubstituted ring seems to interact with the protein and yet its binding causing significant conformational change: the gating loop is further from the substrate-binding position than in any other structure and tryptophan 110 has rotated by 180 degrees, out of the active site.
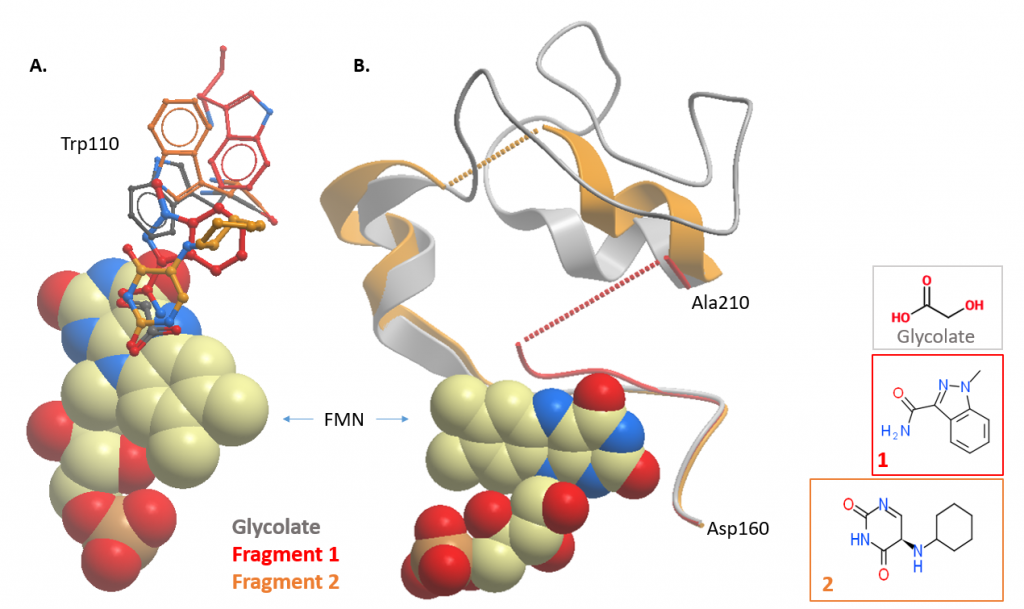
Figure 3: HAO1 structures with active site fragments (fragments 1 and 2; PDB ID 5QIH and 5QIB respectively). Overlay of fragment-bound and glycolate-bound structures showing changes in A. tryptophan 110, and B. the gating loop.
Analysis of gating loop hit
The key lesson here is the discovery of a novel, druggable pocket, bound with fragment 5. This is exciting because inhibitors that target an enzyme’s active site frequently suffer from specificity problems, inhibiting enzymes other than the target, since the active sites of enzymes with similar functions (in this case, flavin-dependent oxidation) are often very similar in structure. Inhibitors targeting this new pocket – which is not conserved in sequence between similar proteins – are much more likely to be specific to HAO1 alone. This is great news for drug development as increasing specificity proportionally decreases off-target drug actions that result in unwanted, sometimes serious, side effects.
Fragments at the subunit interface
Of the two fragments (fragments 6 and 7) at the subunit interface, only fragment 6 was sufficiently buried between the two subunits to be promising for further development. Given the small size of the fragment, it would be unlikely to disrupt the protein tetramer (and so HAO1 activity). Rather, fragment 6 will need to be grown to confer inhibitory activity. Another complication is that tetramer units contribute to the crystal arrangement of HAO1 and so highly ordered crystals are likely to be incompatible with compounds that effectively disrupt tetramer formation i.e. compounds either will not bind or will damage the crystal upon binding. This means that an alternative method will be required to analyse the binding modes of successful hits at this site.
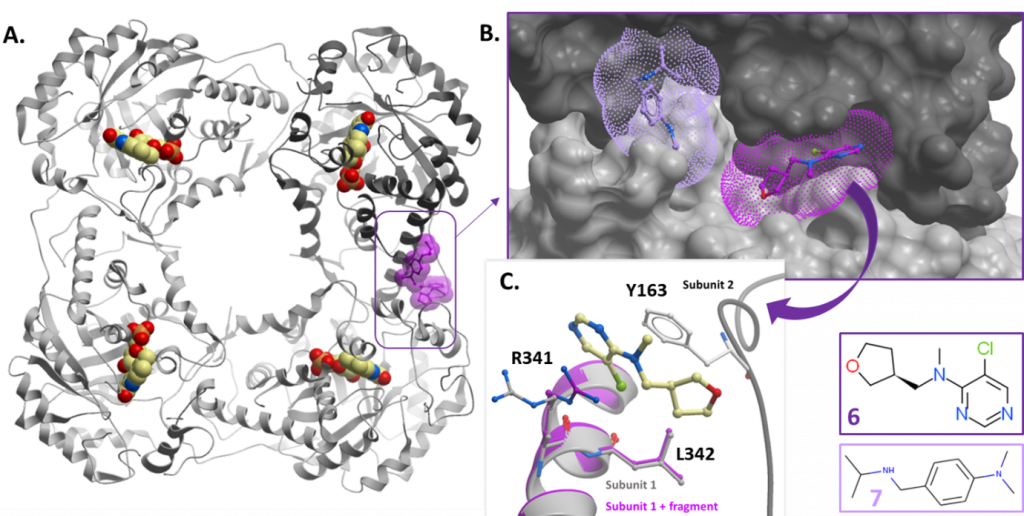
Figure 4: Fragment hits at the subunit interface. A. Position of hits relative to tetramer subunits – fragments shown as purple space-fill models. Fragment 6 (PDB ID 5QID) is in dark purple and fragment 7 (PDB ID 5QIE) in light purple. B. Surface representation of subunit interface showing fragment binding pockets. C. Close up showing coordinating residues for fragment 6.
In my next post, I will tell you about the assays I have used to characterise how the fragments behave towards HAO1 in solution.

One Reply to “Analysis of fragments that bind to HAO1”