As describe in my previous post, my goal is to discover non-catalytic druggable pockets in human enzymes. These pockets could potentially be exploited for the design of ProxPharm compounds (chimeric compounds that bring two proteins in close proximity to elicit an effect of one protein on the other1). An essential aspect for the design of ProxPharm compounds is that the chemical moiety binding the enzyme binds to a non-catalytic pocket, because the compound should not inhibit the activity of the recruited enzyme.
Previously, the methods of both Jiayan Wang’s2 and Setayesh Yazdani’s3 project were used to identify the pockets in all human proteins available in PDB regardless if they were bound to small-molecule ligands. For this, the icmPocketFinder module was used in ICM software (Molsoft, San Diego). In my previous post, I categorized pockets as either catalytic or non-catalytic by measuring the distance between the pocket and catalytic residues present in the structure. The catalytic residues were obtained from the Mechanism and Catalytic Site Atlas (M-CSA) database4 or the UniprotKB databse5 (Figure 1 (Green))6.
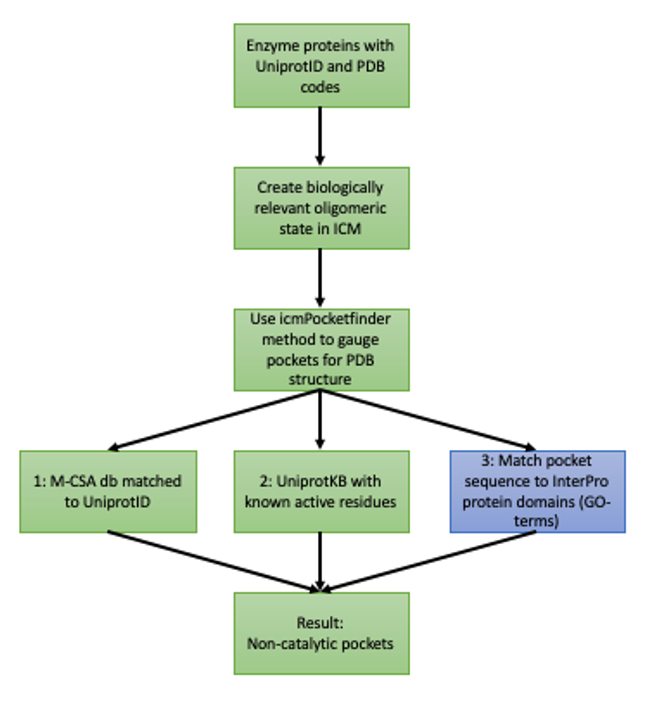
Figure 1. The workflow to distinguish between catalytic and non-catalytic pockets. Green boxes represent first approach (first post) and blue boxes extended second approach.
This method yielded a low amount (1478) of human enzymes to be analyzed, because both databases contained limited information on catalytic residues for human enzymes. Therefore, a second approach was tested, namely identifying whether a pocket is in a catalytic domain by (1) matching the domain information from the InterPro database7 to the pocket and (2) using domain-specific ontology annotations to define the domain as catalytic or not. First, residues within 2.8 Å from the identified pocket were defined as pocket residues. Pocket residues were then associated to protein domains. Based on the GO terms8 of the protein domain, the domain was denoted catalytic or non-catalytic. Only the pockets of UniprotID’s where at least one domain was marked as catalytic were retained for further analysis. Lastly, the pockets flagged as non-catalytic were separated from the pockets that were catalytic and then filtered for distance to catalytic residues (>7 Å) or missing residues (>5 Å) (Final table non_cat_pockets).
Another update in the methodology from the previous post is that the icmPocketfinder module is no longer applied to individual peptide chains, but rather to the complex. The reason for this change is that upon analyzing peptide chains individually, pockets were found that were at the protein-protein interface of a complex (for example, dimer interactions). Compounds occupying these pockets would disrupt the protein function, which we want to avoid.
In total 178,022 pockets could be identified for 23,621 structures representing 2310 proteins. The final table consisted of 71,592 non-catalytic pockets in 11,367 structures representing 1824 proteins. In my next post, I will remove duplicates (identical pockets that were found in different PDB codes of the same protein) and filter the pockets for druggability (volume, hydrophobicity, buriedness and area). If you would like to analyze the results or read the scripts and detailed description of the methods used in this project, please refer to my Zenodo report here.
References:
- Gerry, C. J. & Schreiber, S. L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat. Chem. Biol. 16, 369–378 (2020).
- Wang, J., Yazdani, S., Han, A. & Schapira, M. Structure-based view of the druggable genome. Drug Discov. Today 25, 561–567 (2020).
- Yazdani, S. & Schapira, M. A Gentle Introduction to The Ligandable Genome Project Method 1. (2020) doi:10.5281/ZENODO.3677177.
- Ribeiro, A. J. M. et al. Mechanism and Catalytic Site Atlas (M-CSA): a database of enzyme reaction mechanisms and active sites. Nucleic Acids Res. 46, D618–D623 (2018).
- Consortium, T. U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
- Rovers, E. & Schapira, M. Distinguishing between catalytic and non-catalytic pockets in the ligandable human genome. (2020) doi:10.5281/ZENODO.4294099.
- Hunter, S. et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 37, 211–215 (2009).
- Dimmer, E. C. et al. The Gene Ontology – Providing a Functional Role in Proteomic Studies. Proteomics8, n/a-n/a (2008).
